|
|
@@ -698,7 +698,15 @@ Completely reimplement analysis from scratch as a reproducible workflow
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
Use newly published methods & algorithms not available during the original
|
|
|
- analysis: SICER, csaw, MOFA, ComBat, sva, GREAT, and more
|
|
|
+ analysis: SICER, csaw, MOFA
|
|
|
+\begin_inset CommandInset citation
|
|
|
+LatexCommand cite
|
|
|
+key "Argelaguet2018"
|
|
|
+literal "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+, ComBat, sva, GREAT, and more
|
|
|
\end_layout
|
|
|
|
|
|
\end_deeper
|
|
|
@@ -728,6 +736,20 @@ Focus on what hypotheses were tested, then select figures that show how
|
|
|
\end_inset
|
|
|
|
|
|
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Maybe reorder these sections to do RNA-seq, then ChIP-seq, then combined
|
|
|
+ analyses?
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
@@ -1184,6 +1206,7 @@ status open
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
filename graphics/CD4-csaw/Promoter Peak Distance Profile-PAGE1-CROP.pdf
|
|
|
+ lyxscale 50
|
|
|
width 100col%
|
|
|
groupId colwidth
|
|
|
|
|
|
@@ -1467,12 +1490,783 @@ status collapsed
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Caption Standard
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Comparison of quantification between Salmon with and without Shoal for identical
|
|
|
+ annotation
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+Ultimately selected shoal as quantification, Ensembl as annotation.
|
|
|
+ Why? Running downstream analyses with all quant methods and both annotations
|
|
|
+ showed very little practical difference, so choice was not terribly important.
|
|
|
+ To note in discussion: reproducible workflow made it easy to do this, enabling
|
|
|
+ an informed decision.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+RNA-seq has a large confounding batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/weights-vs-covars-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-seq-weights-vs-covars"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq sample weights, grouped by experimental and technical covariates
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/PCA-no-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-no-batchsub"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/PCA-naive-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-limma-batchsub"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/PCA-combat-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-ComBat-batchsub"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+RNA-seq batch effect can be partially corrected, but still induces uncorrectable
|
|
|
+ biases in downstream analysis
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Figures showing p-value histograms for within-batch and cross-batch contrasts,
|
|
|
+ showing that cross-batch contrasts have attenuated signal, as do comparisons
|
|
|
+ within the bad batch
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+ChIP-seq blacklisting is important
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/csaw/CCF-plots-PAGE2-CROP.pdf
|
|
|
+ lyxscale 50
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Cross-correlation plots with blacklisted reads removed
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/csaw/CCF-plots-noBL-PAGE2-CROP.pdf
|
|
|
+ lyxscale 50
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Cross-correlation plots without removing blacklisted reads
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Un-break the figure legend
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/csaw/CCF-max-plot-CROP.pdf
|
|
|
+ lyxscale 50
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Estimated fragment size in samples before and after blacklisting
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+ChIP-seq normalization
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-sample-MAplot-bins-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+MA plot of H3K4me2 read counts in 10kb bins for two arbitrary samples
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-sample-MAplot-bins-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+MA plot of H3K4me3 read counts in 10kb bins for two arbitrary samples
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K27me3-sample-MAplot-bins-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+MA plot of H3K27me3 read counts in 10kb bins for two arbitrary samples
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+ChIP-seq must be corrected for hidden confounding factors
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-PCA-raw-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me2-bad"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me2 windows, before subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-PCA-SVsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me2-good"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me2 windows, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-PCA-raw-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me3-bad"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me3 windows, before subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-PCA-SVsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me3-good"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me3 windows, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K27me3-PCA-raw-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K27me3-bad"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K27me3 windows, before subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K27me3-PCA-SVsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K27me3-good"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K27me3 windows, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+Figures showing BCV plots with and without SVA for each histone mark.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Can I do supplementary data on a thesis? This is a lot of plots for this
|
|
|
+ section.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+H3K4 and H3K27 promoter methylation has broadly the expected correlation
|
|
|
+ with gene expression
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+H3K4 is correlated with higher expression, and H3K27 is correlated with
|
|
|
+ lower expression genome-wide
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-Comparison of quantification between Salmon with and without Shoal for identical
|
|
|
- annotation
|
|
|
+Grr, gotta find these figures.
|
|
|
+ Maybe in the old analysis?
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1480,33 +2274,31 @@ Comparison of quantification between Salmon with and without Shoal for identical
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
+\begin_layout Itemize
|
|
|
+Figures showing these correlations: box/violin plots of expression distributions
|
|
|
+ with every combination of peak presence/absence in promoter
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Ultimately selected shoal as quantification, Ensembl as annotation.
|
|
|
- Why? Running downstream analyses with all quant methods and both annotations
|
|
|
- showed very little practical difference, so choice was not terribly important.
|
|
|
- To note in discussion: reproducible workflow made it easy to do this, enabling
|
|
|
- an informed decision.
|
|
|
+Appropriate statistical tests showing significant differences in expected
|
|
|
+ directions
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
-RNA-seq has a large confounding batch effect
|
|
|
+MOFA recovers biologically relevant variation from blind analysis by correlating
|
|
|
+ across datasets
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/RNA-seq/weights-vs-covars-CROP.png
|
|
|
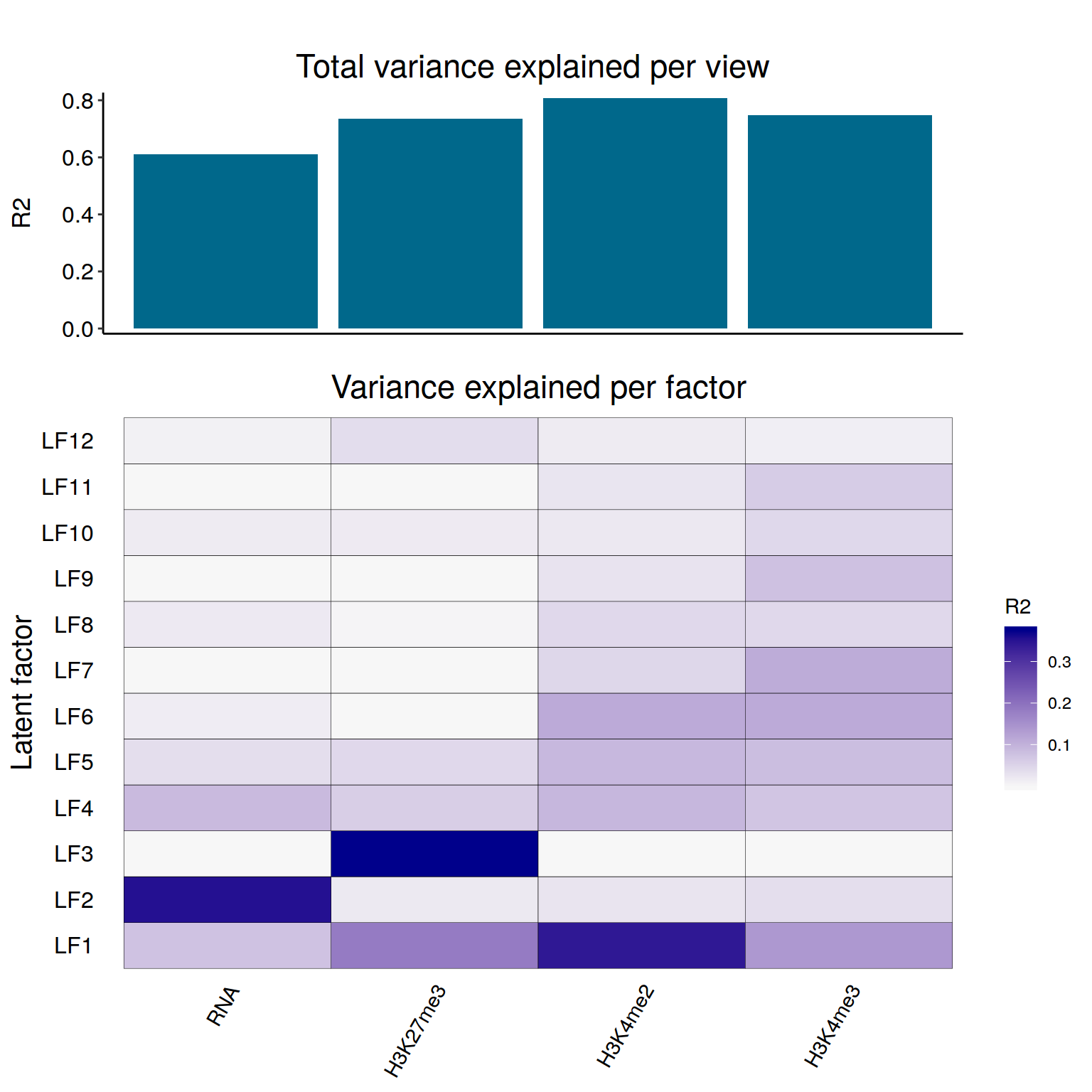
+ filename graphics/CD4-csaw/MOFA-varExplaiend-matrix-CROP.png
|
|
|
lyxscale 25
|
|
|
width 100col%
|
|
|
groupId colwidth-raster
|
|
|
@@ -1524,11 +2316,11 @@ status open
|
|
|
\series bold
|
|
|
\begin_inset CommandInset label
|
|
|
LatexCommand label
|
|
|
-name "fig:RNA-seq-weights-vs-covars"
|
|
|
+name "fig:mofa-varexplained"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq sample weights, grouped by experimental and technical covariates
|
|
|
+Variance explained in each data set by each latent factor estimated by MOFA.
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1536,25 +2328,35 @@ RNA-seq sample weights, grouped by experimental and technical covariates
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Plain Layout
|
|
|
+\end_inset
|
|
|
+
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\end_inset
|
|
|
+\begin_layout Itemize
|
|
|
+Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:mofa-varexplained"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
+\end_inset
|
|
|
|
|
|
+ shows that LF1, 4, and 5 explain substantial var in all data sets
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/RNA-seq/PCA-no-batchsub-CROP.png
|
|
|
+ filename graphics/CD4-csaw/MOFA-LF-distributions-CROP.png
|
|
|
lyxscale 25
|
|
|
width 100col%
|
|
|
groupId colwidth-raster
|
|
|
@@ -1572,11 +2374,11 @@ status open
|
|
|
\series bold
|
|
|
\begin_inset CommandInset label
|
|
|
LatexCommand label
|
|
|
-name "fig:RNA-PCA-no-batchsub"
|
|
|
+name "fig:mofa-lf-dist"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA plot showing clear batch effect
|
|
|
+Sample distribution for each latent factor estimated by MOFA.
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1593,12 +2395,12 @@ RNA-seq PCoA plot showing clear batch effect
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/RNA-seq/PCA-naive-batchsub-CROP.png
|
|
|
+ filename graphics/CD4-csaw/MOFA-LF-scatter-CROP.png
|
|
|
lyxscale 25
|
|
|
width 100col%
|
|
|
groupId colwidth-raster
|
|
|
@@ -1616,11 +2418,16 @@ status open
|
|
|
\series bold
|
|
|
\begin_inset CommandInset label
|
|
|
LatexCommand label
|
|
|
-name "fig:RNA-PCA-no-batchsub-1"
|
|
|
+name "fig:mofa-lf-scatter"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA plot showing clear batch effect
|
|
|
+Scatter plots of specific pairs of MOFA latent factors.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1628,21 +2435,45 @@ RNA-seq PCoA plot showing clear batch effect
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Itemize
|
|
|
+Figures
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:mofa-lf-dist"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ and
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:mofa-lf-scatter"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
\end_inset
|
|
|
|
|
|
+ show that those same 3 LFs, (1, 4, & 5) also correlate best with the experiment
|
|
|
+al factors (cell type & time point)
|
|
|
+\end_layout
|
|
|
|
|
|
+\begin_layout Itemize
|
|
|
+LF2 is clearly the RNA-seq batch effect
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/RNA-seq/PCA-combat-batchsub-CROP.png
|
|
|
+ filename graphics/CD4-csaw/MOFA-batch-correct-CROP.png
|
|
|
lyxscale 25
|
|
|
width 100col%
|
|
|
groupId colwidth-raster
|
|
|
@@ -1660,11 +2491,11 @@ status open
|
|
|
\series bold
|
|
|
\begin_inset CommandInset label
|
|
|
LatexCommand label
|
|
|
-name "fig:RNA-PCA-no-batchsub-2"
|
|
|
+name "fig:mofa-batchsub"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA plot showing clear batch effect
|
|
|
+Result of RNA-seq batch-correction using MOFA latent factors
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1678,120 +2509,120 @@ RNA-seq PCoA plot showing clear batch effect
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-RNA-seq batch effect can be partially corrected, but still induces uncorrectable
|
|
|
- biases in downstream analysis
|
|
|
-\end_layout
|
|
|
+Attempting to remove the effect of LF2 (Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:mofa-batchsub"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
-\begin_deeper
|
|
|
-\begin_layout Itemize
|
|
|
-Figure showing MDS plot before & after ComBat
|
|
|
-\end_layout
|
|
|
+\end_inset
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Figure relating sample weights to batches, cell types, time points, etc.,
|
|
|
- showing that one batch is significantly worse quality
|
|
|
-\end_layout
|
|
|
+) results in batch correction comparable to ComBat (Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:RNA-PCA-ComBat-batchsub"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Figures showing p-value histograms for within-batch and cross-batch contrasts,
|
|
|
- showing that cross-batch contrasts have attenuated signal, as do comparisons
|
|
|
- within the bad batch
|
|
|
-\end_layout
|
|
|
+\end_inset
|
|
|
|
|
|
-\end_deeper
|
|
|
-\begin_layout Subsection
|
|
|
-ChIP-seq must be corrected for hidden confounding factors
|
|
|
+)
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Figures showing pre- and post-SVA MDS plots for each histone mark
|
|
|
+MOFA was able to do this batch subtraction without directly using the sample
|
|
|
+ labels (sample labels were used implicitly to select which factor to subtract)
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Figures showing BCV plots with and without SVA for each histone mark
|
|
|
+Similarity of results shows that batch correction can't get much better
|
|
|
+ than ComBat (despite ComBat ignoring time point)
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
-H3K4 and H3K27 promoter methylation has broadly the expected correlation
|
|
|
- with gene expression
|
|
|
+Naive-to-memory convergence observed in H3K4 and RNA-seq data, not in H3K27me3
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-H3K4 is correlated with higher expression, and H3K27 is correlated with
|
|
|
- lower expression genome-wide
|
|
|
-\end_layout
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Figures showing these correlations: box/violin plots of expression distributions
|
|
|
- with every combination of peak presence/absence in promoter
|
|
|
-\end_layout
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/PCA-final-23-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Appropriate statistical tests showing significant differences in expected
|
|
|
- directions
|
|
|
-\end_layout
|
|
|
|
|
|
-\begin_layout Subsection
|
|
|
-MOFA recovers biologically relevant variation from blind analysis by correlating
|
|
|
- across datasets
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-MOFA
|
|
|
-\begin_inset CommandInset citation
|
|
|
-LatexCommand cite
|
|
|
-key "Argelaguet2018"
|
|
|
-literal "false"
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-group"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- successfully separates biologically relevant patterns of variation from
|
|
|
- technical confounding factors without knowing the sample labels, by finding
|
|
|
- latent factors that explain variation across multiple data sets.
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_deeper
|
|
|
-\begin_layout Itemize
|
|
|
-Figure: show percent-variance-explained plot from MOFA and PCA-like plots
|
|
|
- for the relevant latent factors
|
|
|
-\end_layout
|
|
|
+\end_inset
|
|
|
+
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-MOFA analysis also shows that batch effect correction can't get much better
|
|
|
- than it already is (Figure comparing blind MOFA batch correction to ComBat
|
|
|
- correction)
|
|
|
\end_layout
|
|
|
|
|
|
-\end_deeper
|
|
|
-\begin_layout Subsection
|
|
|
-Naive-to-memory convergence observed in H3K4 and RNA-seq data, not in H3K27me3
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
H3K4 and RNA-seq data show clear evidence of naive convergence with memory
|
|
|
- between days 1 and 5 (MDS plot figure, also compare with last figure from
|
|
|
-
|
|
|
-\begin_inset CommandInset citation
|
|
|
-LatexCommand cite
|
|
|
-key "LaMere2016"
|
|
|
-literal "false"
|
|
|
+ between days 1 and 5 (Figures
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:PCoA-H3K4me2-good"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-)
|
|
|
-\end_layout
|
|
|
+,
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:PCoA-H3K4me3-good"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
+\end_inset
|
|
|
|
|
|
-\begin_layout Plain Layout
|
|
|
-Note that Sarah has granted permission to use her figures
|
|
|
-\end_layout
|
|
|
+, and
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:RNA-PCA-group"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-
|
|
|
+.
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
@@ -1800,7 +2631,17 @@ Table of numbers of genes different between N & M at each time point, showing
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Similar figure for H3K27me3 showing lack of convergence
|
|
|
+Similar figure for H3K27me3 showing lack of convergence (Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:PCoA-H3K27me3-good"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+)
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
@@ -1841,6 +2682,12 @@ MOFA shows great promise for accelerating discovery of major biological
|
|
|
\end_layout
|
|
|
|
|
|
\begin_deeper
|
|
|
+\begin_layout Itemize
|
|
|
+MOFA successfully separates biologically relevant patterns of variation
|
|
|
+ from technical confounding factors without knowing the sample labels, by
|
|
|
+ finding latent factors that explain variation across multiple data sets.
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
MOFA was added to this analysis late and played primarily a confirmatory
|
|
|
role, but it was able to confirm earlier conclusions with much less prior
|
|
|
@@ -1860,9 +2707,59 @@ Naive-to-memory convergence implies that naive cells are differentiating
|
|
|
in this differentiation while H3K27me3 is less involved
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/LaMere2016_fig8.pdf
|
|
|
+ lyxscale 50
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+LaMere 2016 Figure 8, reproduced with permission.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+Convergence is consistent with Lamere2016 fig 8
|
|
|
+\begin_inset CommandInset citation
|
|
|
+LatexCommand cite
|
|
|
+key "LaMere2016"
|
|
|
+literal "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ (which was created without the benefit of SVA)
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
H3K27me3, canonically regarded as a deactivating mark, seems to have a more
|
|
|
- complex
|
|
|
+ complex effect
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
@@ -2894,6 +3791,7 @@ status collapsed
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
filename graphics/PAM/predplot.pdf
|
|
|
+ lyxscale 50
|
|
|
width 100col%
|
|
|
groupId colwidth
|
|
|
|
|
|
@@ -2974,6 +3872,7 @@ status collapsed
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
filename graphics/PAM/ROC-TXvsAR-internal.pdf
|
|
|
+ lyxscale 50
|
|
|
width 100col%
|
|
|
groupId colwidth
|
|
|
|
|
|
@@ -3632,6 +4531,7 @@ status collapsed
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
filename graphics/PAM/ROC-TXvsAR-external.pdf
|
|
|
+ lyxscale 50
|
|
|
width 100col%
|
|
|
groupId colwidth
|
|
|
|