|
|
@@ -1246,7 +1246,7 @@ literal "false"
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
-RNA-seq align+quant method selection
|
|
|
+RNA-seq align+quant method comparison
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
@@ -1254,7 +1254,7 @@ RNA-seq align+quant method selection
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-Maybe fix up the axis ranges for these plots?
|
|
|
+Maybe fix up the excessive axis ranges for these plots?
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1415,7 +1415,7 @@ Comparison of quantification between STAR and Salmon for identical annotation
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1452,7 +1452,7 @@ n
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status open
|
|
|
+status collapsed
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1485,14 +1485,198 @@ Comparison of quantification between Salmon with and without Shoal for identical
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Standard
|
|
|
-
|
|
|
+\begin_layout Itemize
|
|
|
+Ultimately selected shoal as quantification, Ensembl as annotation.
|
|
|
+ Why? Running downstream analyses with all quant methods and both annotations
|
|
|
+ showed very little practical difference, so choice was not terribly important.
|
|
|
+ To note in discussion: reproducible workflow made it easy to do this, enabling
|
|
|
+ an informed decision.
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
RNA-seq has a large confounding batch effect
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
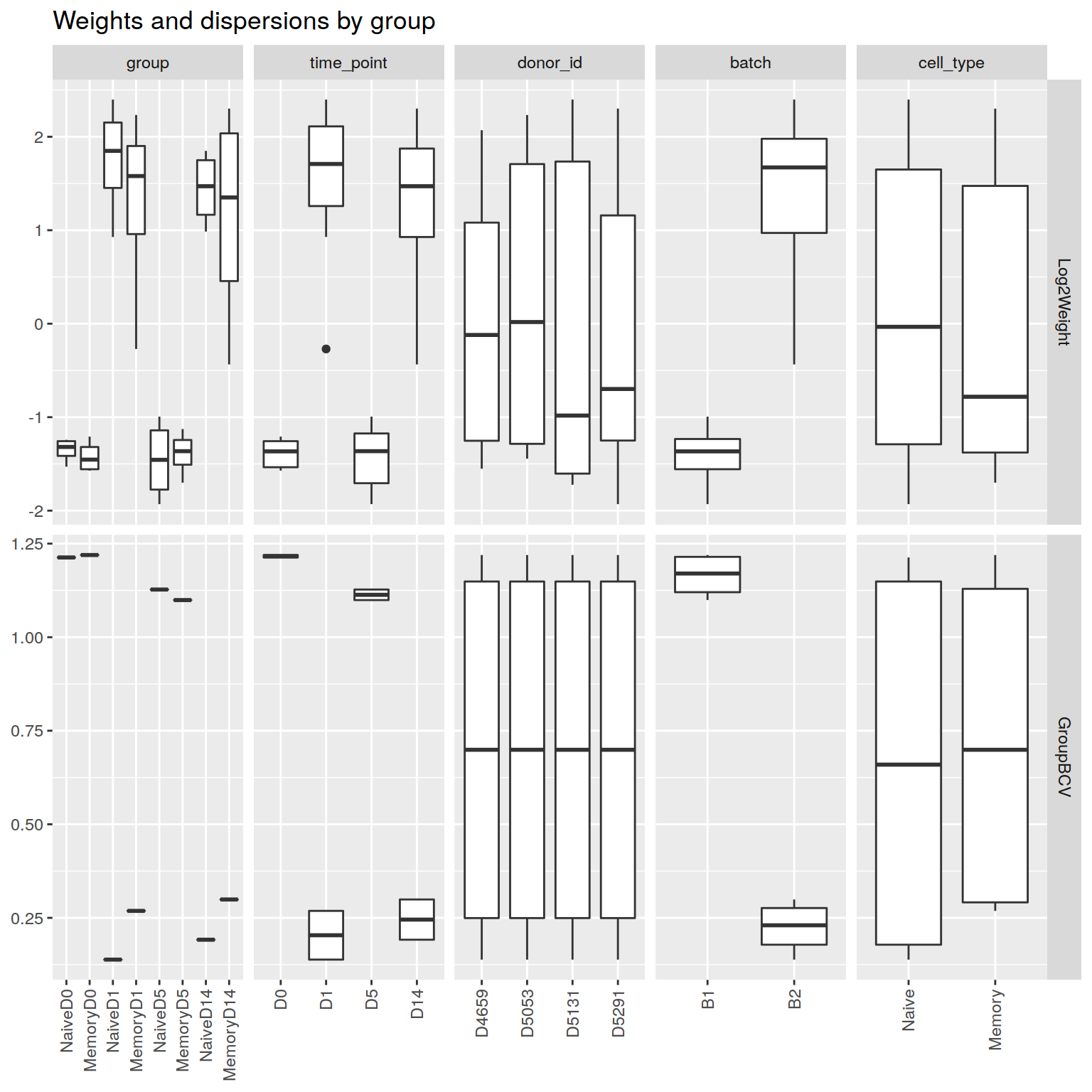
+ filename graphics/CD4-csaw/RNA-seq/weights-vs-covars-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-seq-weights-vs-covars"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq sample weights, grouped by experimental and technical covariates
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
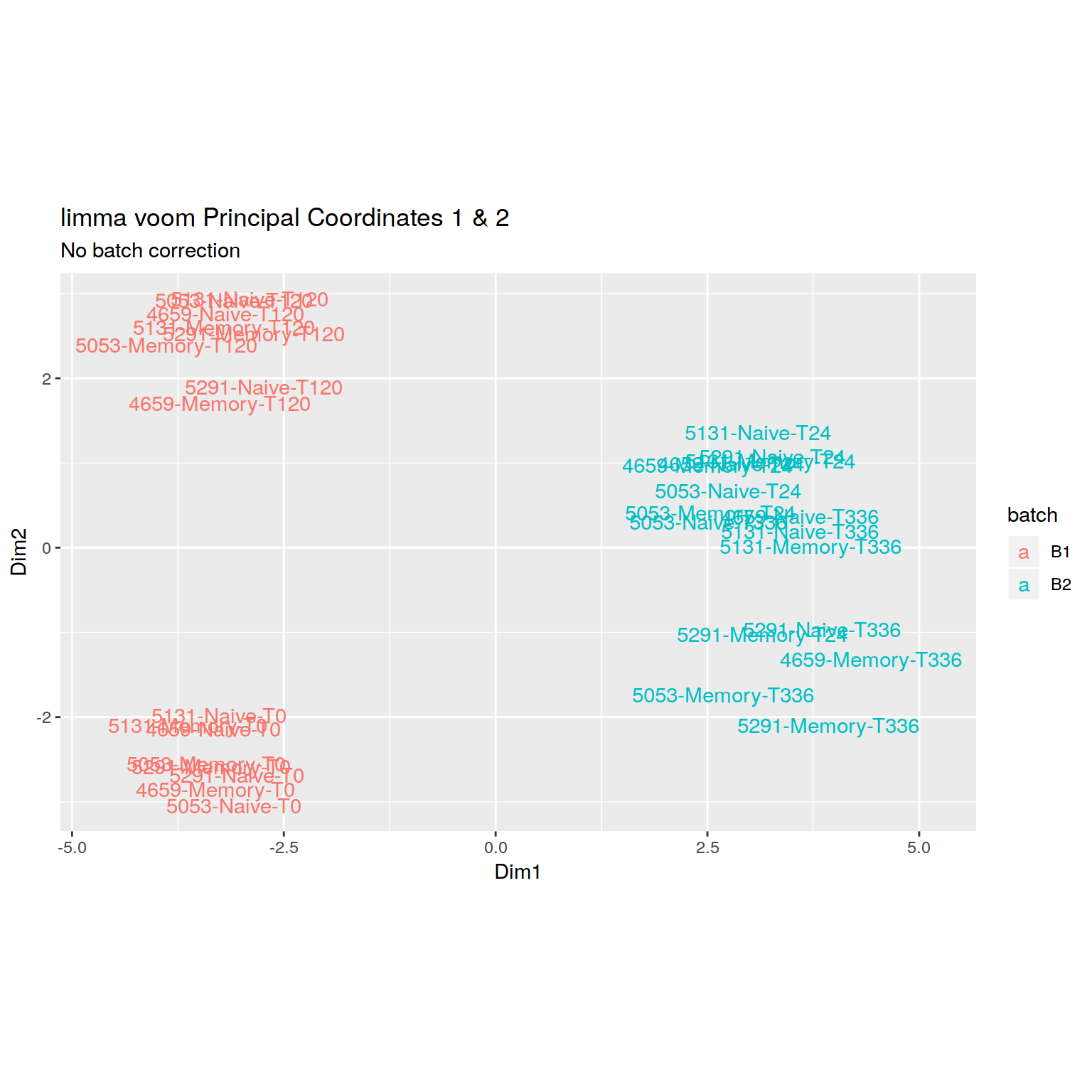
+ filename graphics/CD4-csaw/RNA-seq/PCA-no-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-no-batchsub"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
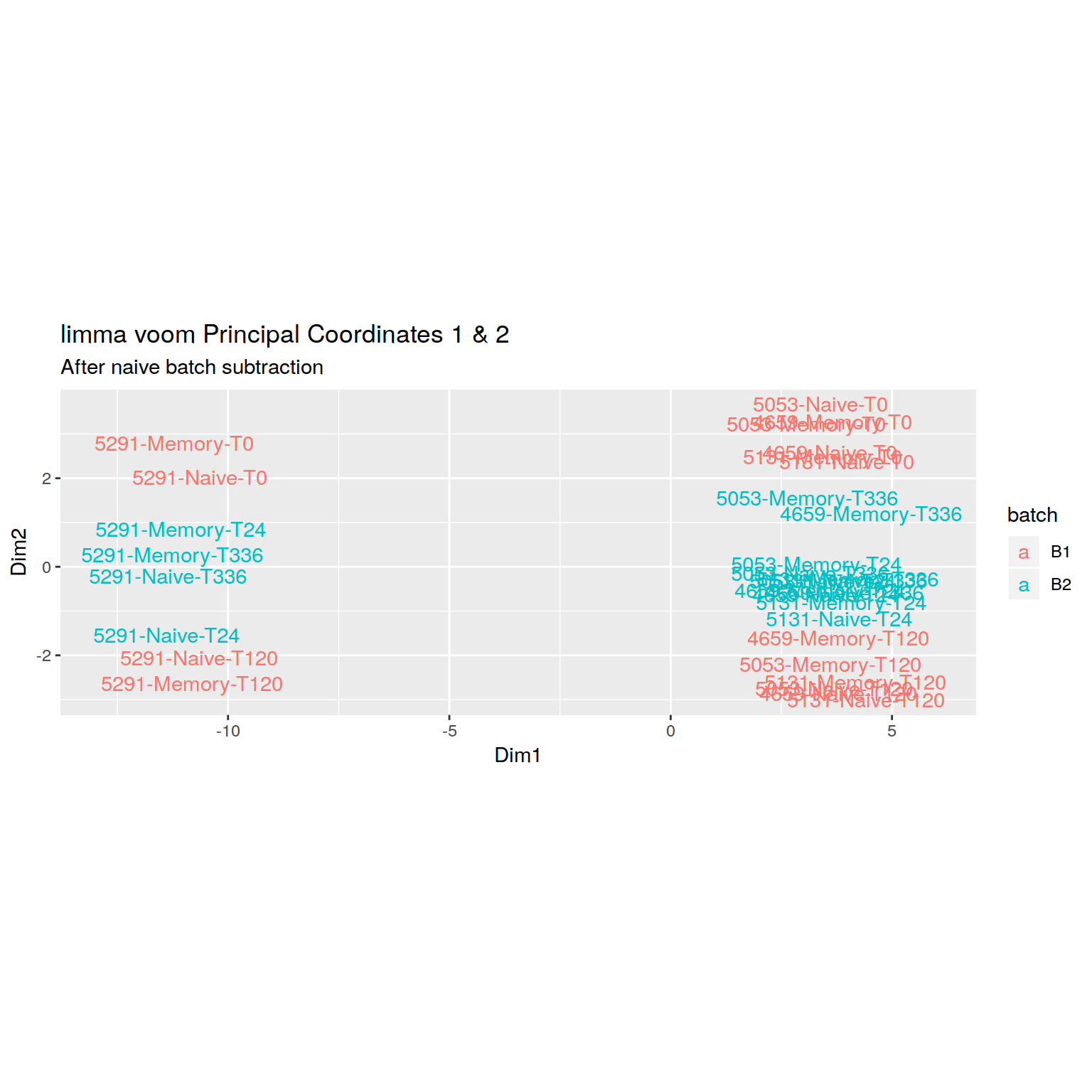
+ filename graphics/CD4-csaw/RNA-seq/PCA-naive-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-no-batchsub-1"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
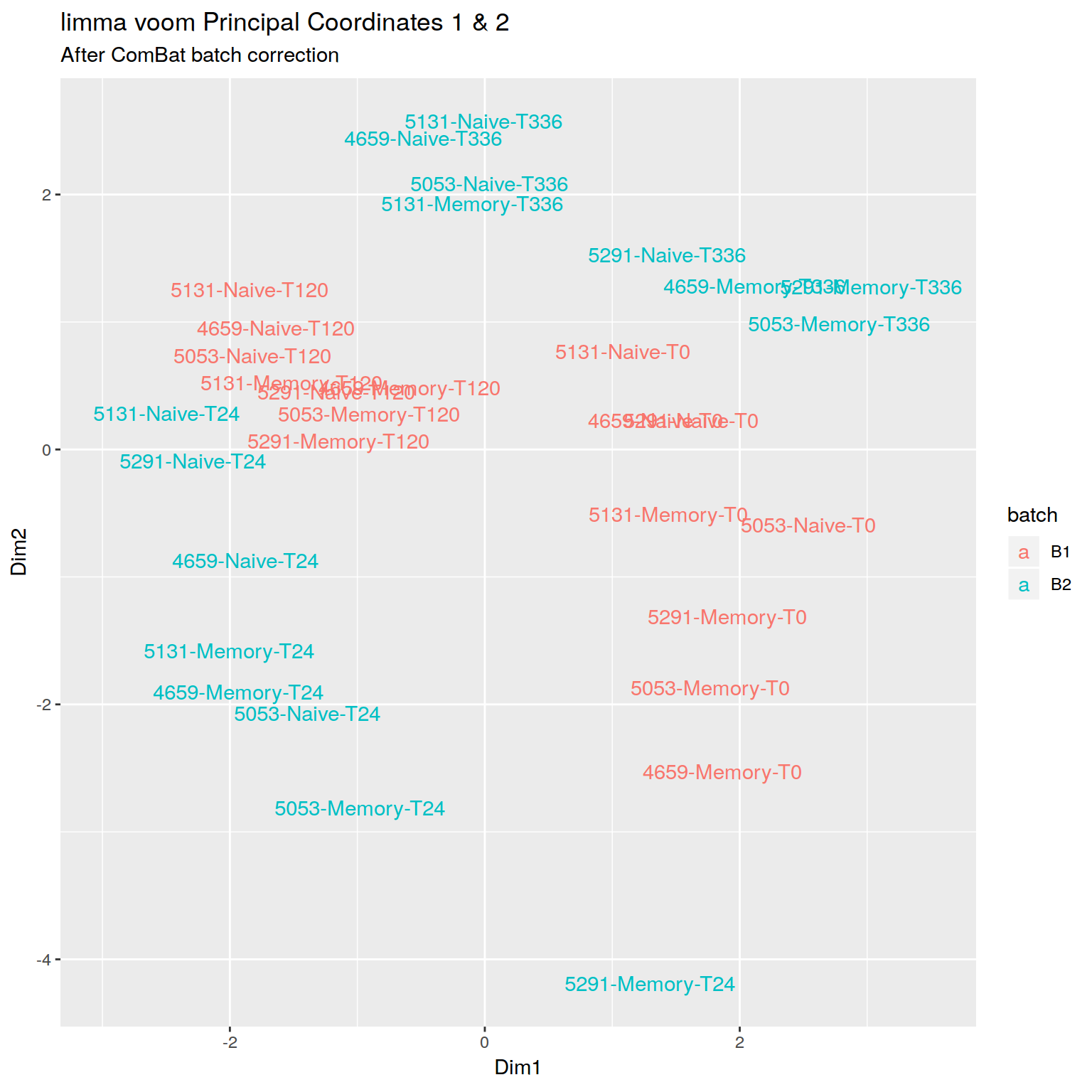
+ filename graphics/CD4-csaw/RNA-seq/PCA-combat-batchsub-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-no-batchsub-2"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA plot showing clear batch effect
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
RNA-seq batch effect can be partially corrected, but still induces uncorrectable
|
|
|
biases in downstream analysis
|