|
|
@@ -5899,20 +5899,6 @@ trajectory
|
|
|
\end_inset
|
|
|
|
|
|
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-Which promoters were considered? Only ones with peaks? Only expressed genes?
|
|
|
- I don't recall exactly the filtering criteria.
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
@@ -5957,7 +5943,7 @@ TSS
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- with 21 windows.
|
|
|
+ with a total of 21 windows.
|
|
|
Reads in each window for each
|
|
|
\begin_inset Flex Glossary Term
|
|
|
status open
|
|
|
@@ -5980,6 +5966,47 @@ logCPM
|
|
|
\end_inset
|
|
|
|
|
|
as in the differential modification analysis.
|
|
|
+ An abundance threshold was chosen such that 99% of peak-containing promoters
|
|
|
+ have an average
|
|
|
+\begin_inset Flex Glossary Term
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+logCPM
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ above this threshold (Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:Promoter-abundance-filtering"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+).
|
|
|
+ Then
|
|
|
+\emph on
|
|
|
+all
|
|
|
+\emph default
|
|
|
+promoters with an average
|
|
|
+\begin_inset Flex Glossary Term
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+logCPM
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ above this threshold were included, and all below that thereshold were
|
|
|
+ filtered out, regardless of whether they actually contained a called peak.
|
|
|
+ This ensures that even promoters containing undetected peaks will be included,
|
|
|
+ at the cost of likely including many promoters that do not contain any
|
|
|
+ true peak.
|
|
|
Then, the
|
|
|
\begin_inset Flex Glossary Term
|
|
|
status open
|
|
|
@@ -5990,9 +6017,9 @@ logCPM
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- values within each promoter were normalized to an average of zero, such
|
|
|
- that each window's normalized abundance now represents the relative read
|
|
|
- depth of that window compared to all other windows in the same promoter.
|
|
|
+ values of the bins within each promoter were normalized to an average of
|
|
|
+ zero, such that each window's normalized abundance now represents the relative
|
|
|
+ read depth of that window compared to all other windows in the same promoter.
|
|
|
The normalized abundance values for each window in a promoter are collectively
|
|
|
referred to as that promoter's
|
|
|
\begin_inset Quotes eld
|
|
|
@@ -6005,6 +6032,214 @@ relative coverage profile
|
|
|
.
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset ERT
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+
|
|
|
+\backslash
|
|
|
+afterpage{
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+
|
|
|
+\backslash
|
|
|
+begin{landscape}
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-neighborhood-logCPM-filter.png
|
|
|
+ lyxscale 25
|
|
|
+ width 30col%
|
|
|
+ groupId nhood-filter-subfig
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me2
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\begin_inset space \hfill{}
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-neighborhood-logCPM-filter.png
|
|
|
+ lyxscale 25
|
|
|
+ width 30col%
|
|
|
+ groupId nhood-filter-subfig
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\begin_inset space \hfill{}
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K27me3-neighborhood-logCPM-filter.png
|
|
|
+ lyxscale 25
|
|
|
+ width 30col%
|
|
|
+ groupId nhood-filter-subfig
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K27me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Argument 1
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
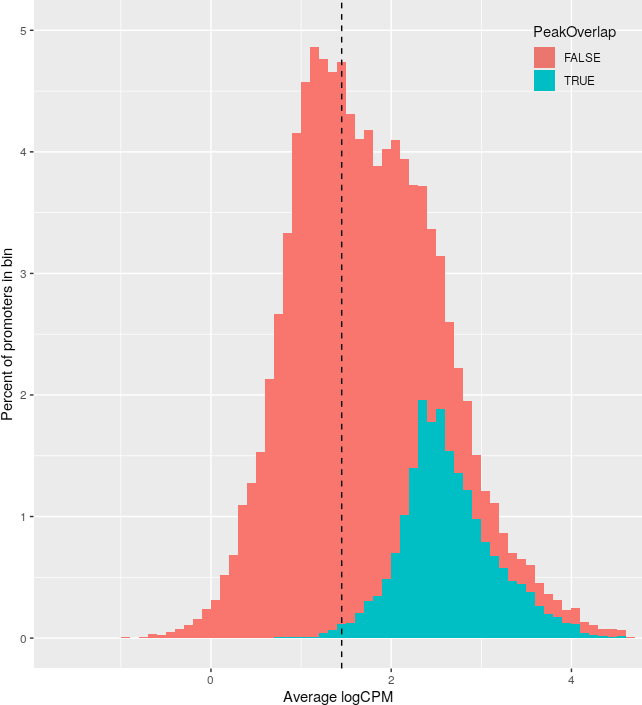
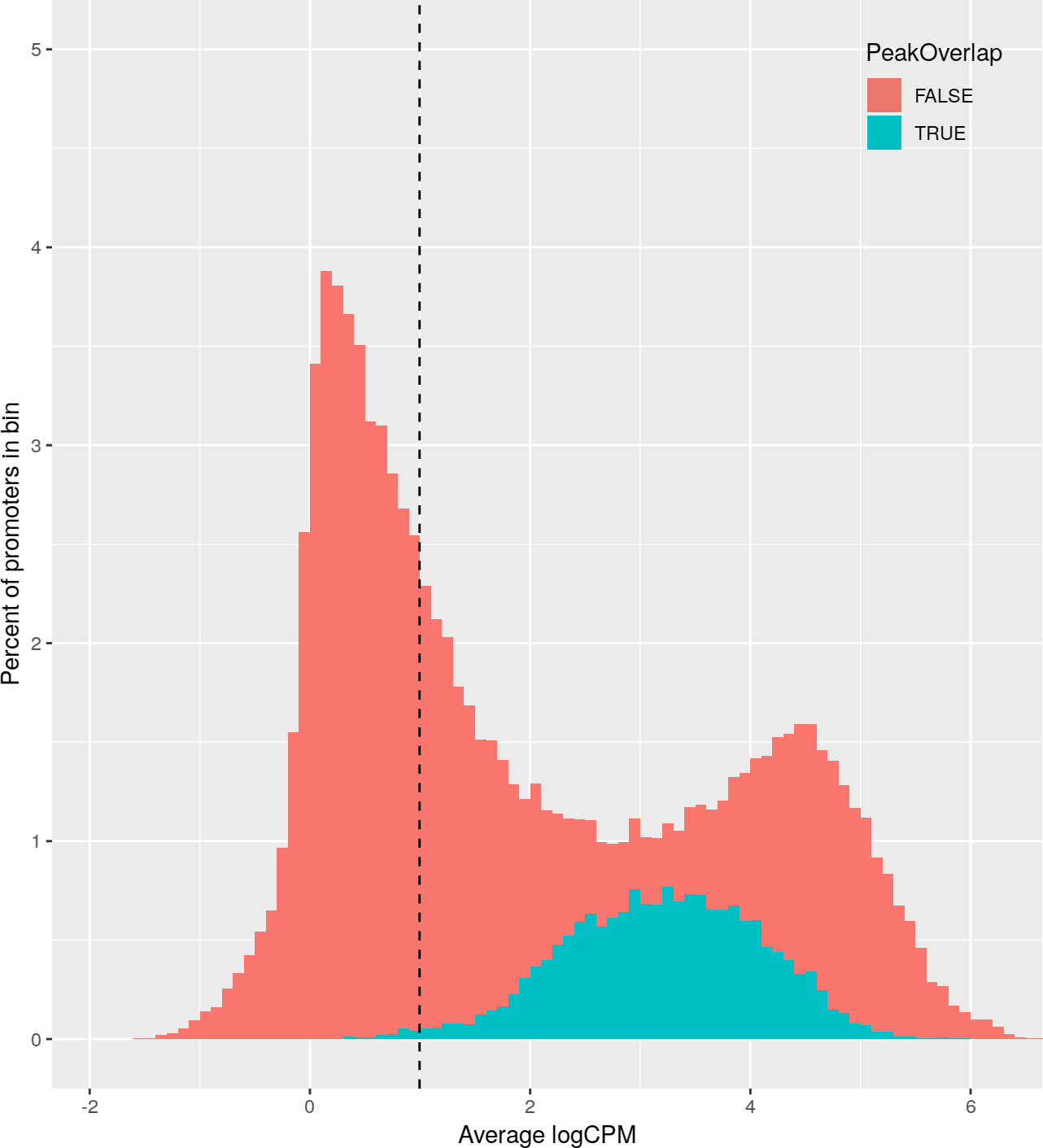
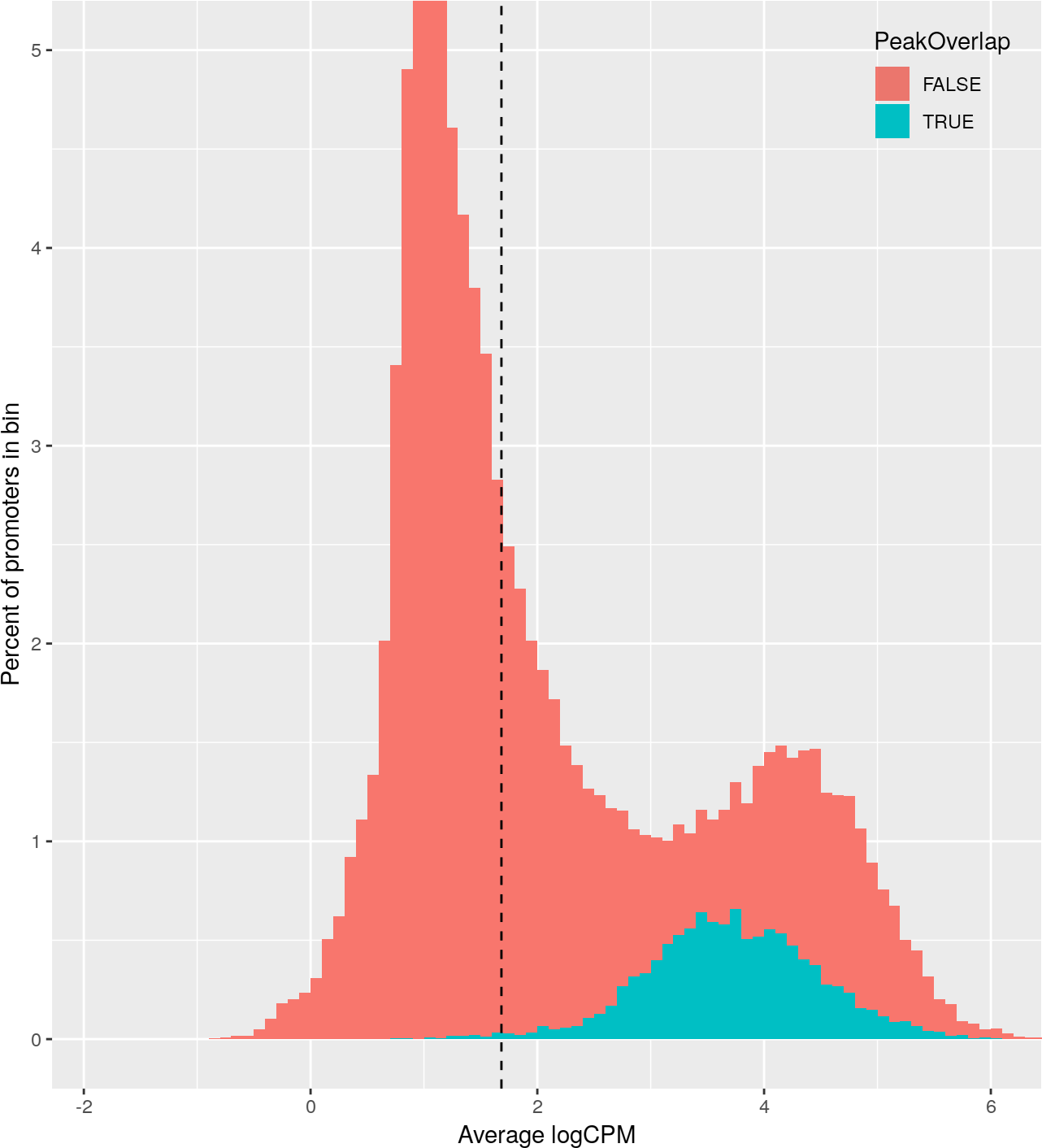
+Promoter abundance filtering for relative coverage profiles.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:Promoter-abundance-filtering"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\series bold
|
|
|
+Promoter abundance filtering for relative coverage profiles.
|
|
|
+
|
|
|
+\series default
|
|
|
+ For each histone mark, a histogram of promoter logCPM values was plotted,
|
|
|
+ colored by whether each promoter contains a called peak.
|
|
|
+ The abundance filter for each histone mark (dotted vertical line) was set
|
|
|
+ such that 99% of peak-containing promoters (blue) are above the threshold,
|
|
|
+ and then all promoters above this threshold were included in downstream
|
|
|
+ analyses.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset ERT
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+
|
|
|
+\backslash
|
|
|
+end{landscape}
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+}
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Subsection
|
|
|
MOFA analysis of cross-dataset variation patterns
|
|
|
\end_layout
|