|
|
@@ -237,6 +237,23 @@ Data generated by Sarah Lamere, published in GEO as
|
|
|
|
|
|

|
|
|
|
|
|
+## Questions to focus on
|
|
|
+
|
|
|
+::: incremental
|
|
|
+
|
|
|
+1. How do we define the "promoter region" for each gene? \vspace{10pt}
|
|
|
+2. How do these histone marks behave in promoter regions? \vspace{10pt}
|
|
|
+3. What can these histone marks tell us about T-cell activation and
|
|
|
+ differentiation?
|
|
|
+
|
|
|
+:::
|
|
|
+
|
|
|
+## First question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+How do we define the "promoter region" for each gene?
|
|
|
+
|
|
|
## Histone modifications occur on consecutive histones
|
|
|
|
|
|
![ChIP-seq coverage in IL2 gene[^lamerethesis]](graphics/presentation/LaMere-thesis-fig3.9-SVG-CROP.png){ height=65% }
|
|
|
@@ -271,7 +288,7 @@ Data generated by Sarah Lamere, published in GEO as
|
|
|
|
|
|
## Each histone mark has an "effective promoter radius"
|
|
|
|
|
|
-
|
|
|
+
|
|
|
|
|
|
## Peaks in promoters correlate with gene expression
|
|
|
|
|
|
@@ -285,25 +302,50 @@ Data generated by Sarah Lamere, published in GEO as
|
|
|
\caption{Expression distributions of genes with and without promoter peaks}
|
|
|
\end{figure}
|
|
|
|
|
|
-## The story so far
|
|
|
+## First question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+How do we define the "promoter region" for each gene?
|
|
|
+
|
|
|
+## Answer: Define the promoter region empirically!
|
|
|
|
|
|
<!-- TODO: Left column: text; right column: flip through relevant image -->
|
|
|
|
|
|
-* H3K4me2, H3K4me3, and H3K27me3 occur on many consecutive histones in
|
|
|
- broad regions across the genome
|
|
|
-* These enriched regions occur more commonly within a certain radius
|
|
|
- of gene promoters
|
|
|
-* This "effective promoter radius" is consistent across all samples
|
|
|
- for a given histone mark, but differs between histone marks
|
|
|
+:::::::::: {.columns}
|
|
|
+::: {.column width="50%"}
|
|
|
+
|
|
|
+* H3K4me2, H3K4me3, and H3K27me3 occur in broad regions across the
|
|
|
+ genome
|
|
|
+* Enriched regions occur more commonly near promoters
|
|
|
+* Each histone mark has its own "effective promoter radius"
|
|
|
* Presence or absence of a peak within this radius is correlated with
|
|
|
gene expression
|
|
|
-
|
|
|
-. . .
|
|
|
|
|
|
-Next: Does the position of a histone modification within a gene
|
|
|
-promoter matter to that gene's expression, or is it merely the
|
|
|
-presence or absence anywhere within the promoter?
|
|
|
-
|
|
|
+:::
|
|
|
+
|
|
|
+::: {.column width="50%"}
|
|
|
+\centering
|
|
|
+\only<1>{\includegraphics[width=\textwidth,height=0.7\textheight]{graphics/presentation/CCF-plots-A-SVG.png}}
|
|
|
+\only<2>{\includegraphics[width=\textwidth,height=0.7\textheight]{graphics/presentation/Promoter-Peak-Distance-Profile-SVG.pdf}}
|
|
|
+\only<3>{\includegraphics[width=\textwidth,height=0.7\textheight]{graphics/presentation/FPKM-by-Peak-Violin-Plots-A-SVG.png}}
|
|
|
+:::
|
|
|
+::::::::::
|
|
|
+
|
|
|
+## Next question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+How do these histone marks behave in promoter regions?
|
|
|
+
|
|
|
+::: notes
|
|
|
+
|
|
|
+Does the position of a histone modification within a gene promoter
|
|
|
+matter to that gene's expression, or is it merely the presence or
|
|
|
+absence anywhere within the promoter?
|
|
|
+
|
|
|
+:::
|
|
|
+
|
|
|
## H3K4me2 promoter neighborhood K-means clusters
|
|
|
|
|
|
{ height=70% }
|
|
|
@@ -385,7 +427,13 @@ presence or absence anywhere within the promoter?
|
|
|
:::
|
|
|
::::::::::
|
|
|
|
|
|
-## Summary of promoter relative coverage findings
|
|
|
+## Current question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+How do these histone marks behave in promoter regions?
|
|
|
+
|
|
|
+## Answer: Presence and position both matter!
|
|
|
|
|
|
### H3K4me2 & H3K4me3
|
|
|
|
|
|
@@ -396,19 +444,25 @@ presence or absence anywhere within the promoter?
|
|
|
|
|
|
### H3K27me3
|
|
|
|
|
|
-* Depletion of H3K27me3 at TSS associated with elevated gene
|
|
|
+* Depletion of H3K27me3 at TSS $\Rightarrow$ elevated gene expression
|
|
|
+* Enrichment of H3K27me3 upstream of TSS $\Rightarrow$ *more* elevated
|
|
|
expression
|
|
|
-* Enrichment of H3K27me3 upstream of TSS even more strongly associated
|
|
|
- with elevated expression
|
|
|
-* Other coverage profiles not associated with elevated expression
|
|
|
+* Other coverage profiles: no association
|
|
|
+
|
|
|
+## Last question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+What can these histone marks tell us about T-cell activation and
|
|
|
+differentiation?
|
|
|
|
|
|
## Differential modification disappears by Day 14
|
|
|
|
|
|
-
|
|
|
+
|
|
|
|
|
|
## Differential modification disappears by Day 14
|
|
|
|
|
|
-
|
|
|
+
|
|
|
|
|
|
## Promoter H3K4me2 levels converge at Day 14
|
|
|
|
|
|
@@ -456,21 +510,19 @@ presence or absence anywhere within the promoter?
|
|
|
:::
|
|
|
::::::::::
|
|
|
|
|
|
-<!-- TODO: Intro MOFA, motivate by showing uncorrected PCA -->
|
|
|
-
|
|
|
-## MOFA identifies cross-dataset patterns of variation
|
|
|
+## MOFA: cross-dataset factor analysis
|
|
|
|
|
|
-![MOFA factor analysis schematic[^mofa]](graphics/presentation/MOFA-fig1A-SVG.png)
|
|
|
+![MOFA factor analysis schematic[^mofa]](graphics/presentation/MOFA-fig1A-SVG.png){ height=70% }
|
|
|
|
|
|
[^mofa]: [Argelaguet, Velten, et. al. (2018)](https://onlinelibrary.wiley.com/doi/abs/10.15252/msb.20178124)
|
|
|
|
|
|
-## MOFA LFs explain variation in all 4 data sets
|
|
|
+## Some factors are shared while others are not
|
|
|
|
|
|
\centering
|
|
|
|
|
|
{ height=70% }
|
|
|
|
|
|
-## 3 LFs are shared across all 4 data sets
|
|
|
+## 3 factors are shared across all 4 data sets
|
|
|
|
|
|
\centering
|
|
|
|
|
|
@@ -478,17 +530,25 @@ presence or absence anywhere within the promoter?
|
|
|
|
|
|
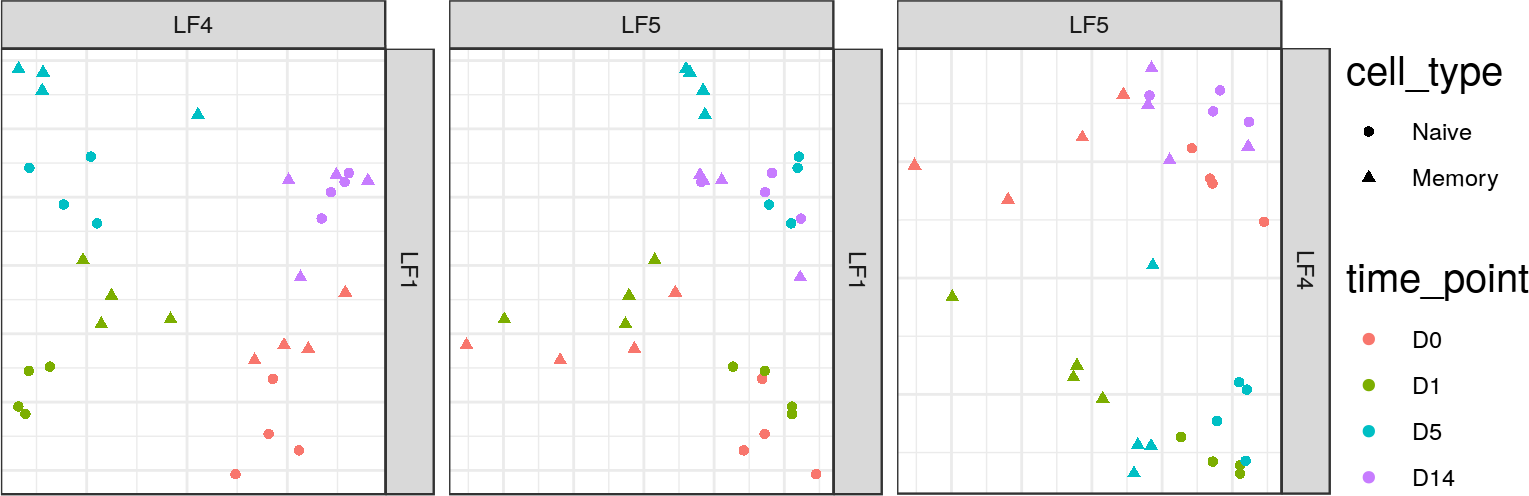
## MOFA LF5 captures convergence pattern
|
|
|
|
|
|
-{ height=70% }
|
|
|
+
|
|
|
|
|
|
-## What have we learned?
|
|
|
+<!-- { height=70% } -->
|
|
|
|
|
|
-* Almost no differential modification observed between naïve and
|
|
|
+## Last question
|
|
|
+
|
|
|
+\centering \LARGE
|
|
|
+
|
|
|
+What can these histone marks tell us about T-cell activation and
|
|
|
+differentiation?
|
|
|
+
|
|
|
+## Answer: Epigenetic convergence between naïve and memory!
|
|
|
+
|
|
|
+* Almost no differential histone modification observed between naïve and
|
|
|
memory at Day 14, despite plenty of differential modification at
|
|
|
earlier time points.
|
|
|
-* RNA-seq data and all 3 histone marks' ChIP-seq data all show
|
|
|
- "convergence" between naïve and memory by Day 14 in the first 2 or 3
|
|
|
- principal coordinates.
|
|
|
-* MOFA captures this convergence pattern in one of the latent factors,
|
|
|
+* Expression and 3 histone marks all show "convergence" between naïve
|
|
|
+ and memory by Day 14 in the first 2 or 3 principal coordinates.
|
|
|
+* MOFA captures this convergence pattern in a single latent factor,
|
|
|
indicating that this is a shared pattern across all 4 data sets.
|
|
|
|
|
|
<!-- ## Slide -->
|
|
|
@@ -496,34 +556,86 @@ presence or absence anywhere within the promoter?
|
|
|
<!--  -->
|
|
|
|
|
|
|
|
|
-## Takeaway 1: Each histone mark has an "effective promoter radius"
|
|
|
+## Questions to focus on
|
|
|
|
|
|
-* H3K4me2, H3K4me3, and H3K27me3 ChIP-seq reads are enriched in broad
|
|
|
- regions across the genome, representing areas where the histone
|
|
|
- modification is present
|
|
|
-* These enriched regions occur more commonly within a certain radius
|
|
|
- of gene promoters
|
|
|
-* This "effective promoter radius" is specific to each histone mark
|
|
|
-* Presence or absence of a peak within this radius is correlated with
|
|
|
- gene expression
|
|
|
+### How do we define the "promoter region" for each gene?
|
|
|
+
|
|
|
+Define empirically using peak-to-promoter distances; validate by
|
|
|
+correlation with expression.
|
|
|
+
|
|
|
+. . .
|
|
|
+
|
|
|
+### How do these histone marks behave in promoter regions?
|
|
|
+
|
|
|
+Location matters! Specific coverage patterns correlated with elevated
|
|
|
+expression.
|
|
|
+
|
|
|
+. . .
|
|
|
+
|
|
|
+### What can we learn about T-cell activation and differentiation?
|
|
|
+
|
|
|
+Epigenetic & expression state of naïve and memory converges late after
|
|
|
+activation, consistent with naïve differentiation into memory.
|
|
|
+
|
|
|
+## Further conclusions & future directions
|
|
|
+
|
|
|
+* "Effective promoter region" is a valid concept but "radius"
|
|
|
+ oversimplifies: seek a better definition
|
|
|
+
|
|
|
+* Coverage profiles were only examined in naïve day 0 samples: further
|
|
|
+ analysis could incorporate time and cell type
|
|
|
+
|
|
|
+* Coverage profile normalization induces degeneracy: adapt a better
|
|
|
+ normalization from peak callers like SICER
|
|
|
+
|
|
|
+* Unimodal distribution of promoter coverage profiles is unexpected
|
|
|
+
|
|
|
+## Further conclusions & future directions
|
|
|
+
|
|
|
+* Experiment was not designed to directly test the epigenetic
|
|
|
+ convergence hypothesis: future experiments could include cultured
|
|
|
+ but un-activated controls
|
|
|
+
|
|
|
+* High correlation between H3K4me3 and H3K4me2 is curious given they
|
|
|
+ are mutually exclusive: design experiments to determine the degree
|
|
|
+ of actual co-occurrence
|
|
|
|
|
|
-## Takeaway 2: Peak position within the promoter is important
|
|
|
-
|
|
|
-* H3K4me2 and H3K4me3 peaks are more strongly associated with elevated
|
|
|
- gene expression the closer they are to the TSS, with a slight bias
|
|
|
- toward downstream peaks.
|
|
|
-* H3K27me3 depletion at the TSS and enrichement upstream are both
|
|
|
- associated with elevated expression, while other patterns are not.
|
|
|
-* In all histone marks, position of modification within promoter
|
|
|
- appears to be an important factor in association with gene
|
|
|
- expression
|
|
|
+## Implications for transplant biology
|
|
|
+
|
|
|
+::: incremental
|
|
|
+
|
|
|
+* Epigenetic regulation through histone methylation is surely involved
|
|
|
+ in immune memory
|
|
|
+
|
|
|
+* Can we stop memory cells from forming by perturbing histone
|
|
|
+ methylation?
|
|
|
+
|
|
|
+* Can we disrupt memory cell function during rejection by perturbing
|
|
|
+ histone methylation?
|
|
|
+
|
|
|
+* Can we suggest druggable targets for better immune suppression by
|
|
|
+ looking at epigenetically upregulated genes in memory cells?
|
|
|
+
|
|
|
+:::
|
|
|
+
|
|
|
+## Acknowledgements
|
|
|
+
|
|
|
+* My mentors, past and present: Drs. Terry Gaasterland, Daniel
|
|
|
+ Salomon, and Andrew Su
|
|
|
+
|
|
|
+* My committee: Drs. Nicholas Schork, Ali Torkamani, Michael
|
|
|
+ Petrascheck, and Luc Teyton.
|
|
|
+
|
|
|
+* My many collaborators in the Salomon Lab
|
|
|
+
|
|
|
+* The Scripps Genomics Core
|
|
|
+
|
|
|
+* My parents, John & Chris Thompson
|
|
|
+
|
|
|
+## {.plain}
|
|
|
+
|
|
|
+\centering
|
|
|
|
|
|
-## Takeaway 3: Expression & epigenetic state both converge at Day 14
|
|
|
+\huge
|
|
|
|
|
|
-* At Day 14, almost no differential modification observed between
|
|
|
- naïve and memory cells
|
|
|
-* Naïve and memory converge visually in PCoA plots
|
|
|
-* Convergence is a shared pattern of variation across all 3 histone
|
|
|
- marks and gene expression
|
|
|
-* This is consistent with the hypothesis that the naïve cells have
|
|
|
- differentiated into a more memory-like phenotype by day 14.
|
|
|
+Questions?
|