|
|
@@ -1215,7 +1215,7 @@ ChIP-seq blacklisting is important
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1234,6 +1234,14 @@ status collapsed
|
|
|
\begin_inset Caption Standard
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:CCF-with-blacklist"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
Cross-correlation plots with blacklisted reads removed
|
|
|
\end_layout
|
|
|
|
|
|
@@ -1251,7 +1259,7 @@ Cross-correlation plots with blacklisted reads removed
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1270,6 +1278,14 @@ status collapsed
|
|
|
\begin_inset Caption Standard
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:CCF-without-blacklist"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
Cross-correlation plots without removing blacklisted reads
|
|
|
\end_layout
|
|
|
|
|
|
@@ -1281,6 +1297,199 @@ Cross-correlation plots without removing blacklisted reads
|
|
|
\end_inset
|
|
|
|
|
|
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+ChIP-seq peak calling
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Replace these figures with a single table of # of peaks called at chosen
|
|
|
+ IDR threshold, showing that SICER has more
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:IDR-RC-H3K4me2"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+Irreproducible Discovery Rate consistency plots for H3K4me2
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:IDR-RC-H3K4me3"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+Irreproducible Discovery Rate consistency plots for H3K4me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:IDR-RC-H3K27me3"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+Irreproducible Discovery Rate consistency plots for H3K27me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+Figures
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:IDR-RC-H3K4me2"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+,
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:IDR-RC-H3K4me3"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+, and
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:IDR-RC-H3K27me3"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ show the IDR rank-consistency plots for peaks called in an arbitrarily-chosen
|
|
|
+ pair of donors.
|
|
|
+ For all 3 histone marks, when the peaks for each donor are ranked according
|
|
|
+ to their scores, SICER produces much more reproducible results between
|
|
|
+ donors.
|
|
|
+ This is consistent with SICER's stated goal of identifying broad peaks,
|
|
|
+ in contrast to MACS, which is designed for identifying sharp peaks.
|
|
|
+ Based on this observation, the SICER peak calls were used for all downstream
|
|
|
+ analyses that involved ChIP-seq peaks.
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
@@ -1423,8 +1632,30 @@ ChIP-seq must be corrected for hidden confounding factors
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-Consolidate these into 1 2x3 grid.
|
|
|
+Consolidate figures
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:PCoA-H3K4me2-bad"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ through
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:PCoA-H3K27me3-good"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ into 1 2x3 grid lettered A through F.
|
|
|
For now, just refer to them as if they were a single figure.
|
|
|
+ Also, do the good PCoA plots belong in the results section since they are
|
|
|
+ also used there?
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1480,7 +1711,7 @@ PCoA plot of H3K4me2 windows, before subtracting surrogate variables
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1568,7 +1799,7 @@ PCoA plot of H3K4me3 windows, before subtracting surrogate variables
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1656,7 +1887,7 @@ PCoA plot of H3K27me3 windows, before subtracting surrogate variables
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -1697,7 +1928,7 @@ PCoA plot of H3K27me3 windows, after subtracting surrogate variables
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Figures showing BCV plots with and without SVA for each histone mark.
|
|
|
+Figures showing BCV plots with and without SVA for each histone mark?
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
@@ -2104,168 +2335,22 @@ H3K4 and H3K27 methylation occur in broad regions and are enriched near
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-Replace these figures with a single table of # of peaks called at chosen
|
|
|
- IDR threshold, showing that SICER has more
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Float figure
|
|
|
+\begin_inset Float table
|
|
|
wide false
|
|
|
sideways false
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
\begin_inset Flex TODO Note (inline)
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Caption Standard
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-
|
|
|
-\series bold
|
|
|
-\begin_inset CommandInset label
|
|
|
-LatexCommand label
|
|
|
-name "fig:IDR-RC-H3K4me2"
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-Irreproducible Discovery Rate consistency plots for H3K4me2
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Float figure
|
|
|
-wide false
|
|
|
-sideways false
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Caption Standard
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-
|
|
|
-\series bold
|
|
|
-\begin_inset CommandInset label
|
|
|
-LatexCommand label
|
|
|
-name "fig:IDR-RC-H3K4me3"
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-Irreproducible Discovery Rate consistency plots for H3K4me3
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Float figure
|
|
|
-wide false
|
|
|
-sideways false
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-Re-generate IDR rank consistency plots for SICER and MACS side-by-side
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\begin_inset Caption Standard
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-
|
|
|
-\series bold
|
|
|
-\begin_inset CommandInset label
|
|
|
-LatexCommand label
|
|
|
-name "fig:IDR-RC-H3K27me3"
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-Irreproducible Discovery Rate consistency plots for H3K27me3
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-
|
|
|
-\end_layout
|
|
|
-
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Float table
|
|
|
-wide false
|
|
|
-sideways false
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-\align center
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
-
|
|
|
-\begin_layout Plain Layout
|
|
|
-Need
|
|
|
-\emph on
|
|
|
-median
|
|
|
-\emph default
|
|
|
- peak width, not mean
|
|
|
+Also get
|
|
|
+\emph on
|
|
|
+median
|
|
|
+\emph default
|
|
|
+ peak width and maybe other quantiles (25%, 75%)
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2324,7 +2409,7 @@ genome coverage
|
|
|
\begin_inset Text
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-read coverage
|
|
|
+FRiP
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2490,7 +2575,12 @@ name "tab:peak-calling-summary"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-SICER+IDR peak-calling summary
|
|
|
+Peak-calling summary.
|
|
|
+
|
|
|
+\series default
|
|
|
+For each histone mark, the number of peaks called using SICER at an IDR
|
|
|
+ threshold of ???, the mean width of those peaks, the fraction of the genome
|
|
|
+ covered by peaks, and the fraction of reads in peaks (FRiP).
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2504,56 +2594,49 @@ SICER+IDR peak-calling summary
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
-Figures
|
|
|
-\begin_inset CommandInset ref
|
|
|
-LatexCommand ref
|
|
|
-reference "fig:IDR-RC-H3K4me2"
|
|
|
-plural "false"
|
|
|
-caps "false"
|
|
|
-noprefix "false"
|
|
|
-
|
|
|
-\end_inset
|
|
|
-
|
|
|
-,
|
|
|
+Table
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:IDR-RC-H3K4me3"
|
|
|
+reference "tab:peak-calling-summary"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-, and
|
|
|
+ gives a summary of the peak calling statistics for each histone mark.
|
|
|
+ Consistent with previous observations [CITATION NEEDED], all 3 histone
|
|
|
+ marks occur in broad regions spanning many consecutive nucleosomes, rather
|
|
|
+ than in sharp peaks as would be expected for a transcription factor or
|
|
|
+ other molecule that binds to specific sites.
|
|
|
+ This conclusion is further supported by Figure
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:IDR-RC-H3K27me3"
|
|
|
+reference "fig:CCF-with-blacklist"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- show the IDR rank-consistency plots for peaks called in an arbitrarily-chosen
|
|
|
- pair of donors.
|
|
|
- For all 3 histone marks, when the peaks for each donor are ranked according
|
|
|
- to their scores, SICER produces much more reproducible results between
|
|
|
- donors.
|
|
|
- This is consistent with SICER's stated goal of identifying broad peaks,
|
|
|
- in contrast to MACS, which is designed for identifying sharp peaks.
|
|
|
- Based on this observation, the SICER peak calls were used for all downstream
|
|
|
- analyses that involved ChIP-seq peaks.
|
|
|
- Table
|
|
|
+, in which a clear nucleosome-sized periodicity is visible in the cross-correlat
|
|
|
+ion value for each sample, indicating that each time a given mark is present
|
|
|
+ on one histone, it is also likely to be found on adjacent histones as well.
|
|
|
+ H3K27me3 enrichment in particular is substantially more broad than either
|
|
|
+ H3K4 mark, with a mean peak width of almost 19,000 bp.
|
|
|
+ This is also reflected in the periodicity observed in Figure
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "tab:peak-calling-summary"
|
|
|
+reference "fig:CCF-with-blacklist"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- gives a summary of the peak calling statistics for each histone mark.
|
|
|
+, which remains strong much farther out for H3K27me3 than the other marks,
|
|
|
+ showing H3K27me3 especially tends to be found on long runs of consecutive
|
|
|
+ histones.
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
@@ -2563,12 +2646,12 @@ sideways false
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-\align center
|
|
|
-\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/Promoter Peak Distance Profile-PAGE1-CROP.pdf
|
|
|
- lyxscale 50
|
|
|
- width 100col%
|
|
|
- groupId colwidth
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Ensure this figure uses the peak calls from the new analysis.
|
|
|
+\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
@@ -2576,18 +2659,31 @@ status open
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-\begin_inset Caption Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-
|
|
|
-\series bold
|
|
|
-\begin_inset CommandInset label
|
|
|
-LatexCommand label
|
|
|
-name "fig:effective-promoter-radius"
|
|
|
+Need a control: shuffle all peaks and repeat, N times.
|
|
|
+ Do real vs shuffled control both in a top/bottom arrangement.
|
|
|
+\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-Enrichment of peaks in promoter neighborhoods.
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Consider counting TSS inside peaks as negative number indicating how far
|
|
|
+
|
|
|
+\emph on
|
|
|
+inside
|
|
|
+\emph default
|
|
|
+ the peak the TSS is (i.e.
|
|
|
+ distance to nearest non-peak area).
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2596,7 +2692,21 @@ Enrichment of peaks in promoter neighborhoods.
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+The H3K4 part of this figure is included in
|
|
|
+\begin_inset CommandInset citation
|
|
|
+LatexCommand cite
|
|
|
+key "LaMere2016"
|
|
|
+literal "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
|
|
|
+ as Fig.
|
|
|
+ S2.
|
|
|
+ Do I need to do anything about that?
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2604,94 +2714,928 @@ Enrichment of peaks in promoter neighborhoods.
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Each histone mark is enriched within a certain radius of gene TSS positions,
|
|
|
- but that radius is different for each mark (figure
|
|
|
-\begin_inset CommandInset ref
|
|
|
-LatexCommand ref
|
|
|
-reference "fig:effective-promoter-radius"
|
|
|
-plural "false"
|
|
|
-caps "false"
|
|
|
-noprefix "false"
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/Promoter Peak Distance Profile-PAGE1-CROP.pdf
|
|
|
+ lyxscale 50
|
|
|
+ width 80col%
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-, previously in
|
|
|
-\begin_inset CommandInset citation
|
|
|
-LatexCommand cite
|
|
|
-key "LaMere2016"
|
|
|
-literal "false"
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:near-promoter-peak-enrich"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
- Fig.
|
|
|
- S2)
|
|
|
+Enrichment of peaks in promoter neighborhoods.
|
|
|
+
|
|
|
+\series default
|
|
|
+This plot shows the distribution of distances from each annotated transcription
|
|
|
+ start site in the genome to the nearest called peak.
|
|
|
+ Each line represents one combination of histone mark, cell type, and time
|
|
|
+ point.
|
|
|
+ Distributions are smoothed using kernel density estimation [CITE?].
|
|
|
+ Transcription start sites that occur
|
|
|
+\emph on
|
|
|
+within
|
|
|
+\emph default
|
|
|
+ peaks were excluded from this plot to avoid a large spike at zero that
|
|
|
+ would overshadow the rest of the distribution.
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Subsection
|
|
|
-H3K4 and H3K27 promoter methylation has broadly the expected correlation
|
|
|
- with gene expression
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
+\begin_inset Float table
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-This section can easily be cut, especially if I can't find those plots.
|
|
|
+\align center
|
|
|
+\begin_inset Tabular
|
|
|
+<lyxtabular version="3" rows="4" columns="2">
|
|
|
+<features tabularvalignment="middle">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Histone mark
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Effective promoter radius
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me2
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+1 kb
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+1 kb
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K27me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+2.5 kb
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+</lyxtabular>
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "tab:effective-promoter-radius"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+Effective promoter radius for each histone mark.
|
|
|
+
|
|
|
+\series default
|
|
|
+ These values represent the approximate distance from transcription start
|
|
|
+ site positions within which an excess of peaks are found, as shown in Figure
|
|
|
+
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:near-promoter-peak-enrich"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Problem: the effective promoter radius concept is an interesting result
|
|
|
+ on its own, hence its placement here.
|
|
|
+ However, it is also important in the methods section, which comes first.
|
|
|
+ What do? Refer forward to this section? Move this section to Methods?
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+All 3 histone marks tend to occur more often near promoter regions, as shown
|
|
|
+ in Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:near-promoter-peak-enrich"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+.
|
|
|
+ The majority of each density distribution is flat, representing the background
|
|
|
+ density of peaks genome-wide.
|
|
|
+ Each distribution has a peak near zero, representing an enrichment of peaks
|
|
|
+ close transcription start site (TSS) positions relative to the remainder
|
|
|
+ of the genome.
|
|
|
+ Interestingly, the
|
|
|
+\begin_inset Quotes eld
|
|
|
+\end_inset
|
|
|
+
|
|
|
+radius
|
|
|
+\begin_inset Quotes erd
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ within which this enrichment occurs is not the same for every histone mark
|
|
|
+ (Table
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "tab:effective-promoter-radius"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+).
|
|
|
+ For H3K4me2 and H3K4me3, peaks are most enriched within 1
|
|
|
+\begin_inset space ~
|
|
|
+\end_inset
|
|
|
+
|
|
|
+kbp of TSS positions, while for H3K27me3, enrichment is broader, extending
|
|
|
+ to 2.5
|
|
|
+\begin_inset space ~
|
|
|
+\end_inset
|
|
|
+
|
|
|
+kbp.
|
|
|
+ These
|
|
|
+\begin_inset Quotes eld
|
|
|
+\end_inset
|
|
|
+
|
|
|
+effective promoter radii
|
|
|
+\begin_inset Quotes erd
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ were used for all further promoter-based analyses.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Consider also showing figure for distance to nearest peak center, and reference
|
|
|
+ median peak size once that is known.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+H3K4 and H3K27 promoter methylation has broadly the expected correlation
|
|
|
+ with gene expression
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+This section can easily be cut, especially if I can't find those plots.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+H3K4 is correlated with higher expression, and H3K27 is correlated with
|
|
|
+ lower expression genome-wide
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Grr, gotta find these figures.
|
|
|
+ Maybe in the old analysis? At least one of these plots is definitely in
|
|
|
+ Sarah's paper.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+Figures showing these correlations: box/violin plots of expression distributions
|
|
|
+ with every combination of peak presence/absence in promoter
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Itemize
|
|
|
+Appropriate statistical tests showing significant differences in expected
|
|
|
+ directions
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+RNA-seq and H3K4 methylation patterns in naive and memory show convergence
|
|
|
+ at day 14
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
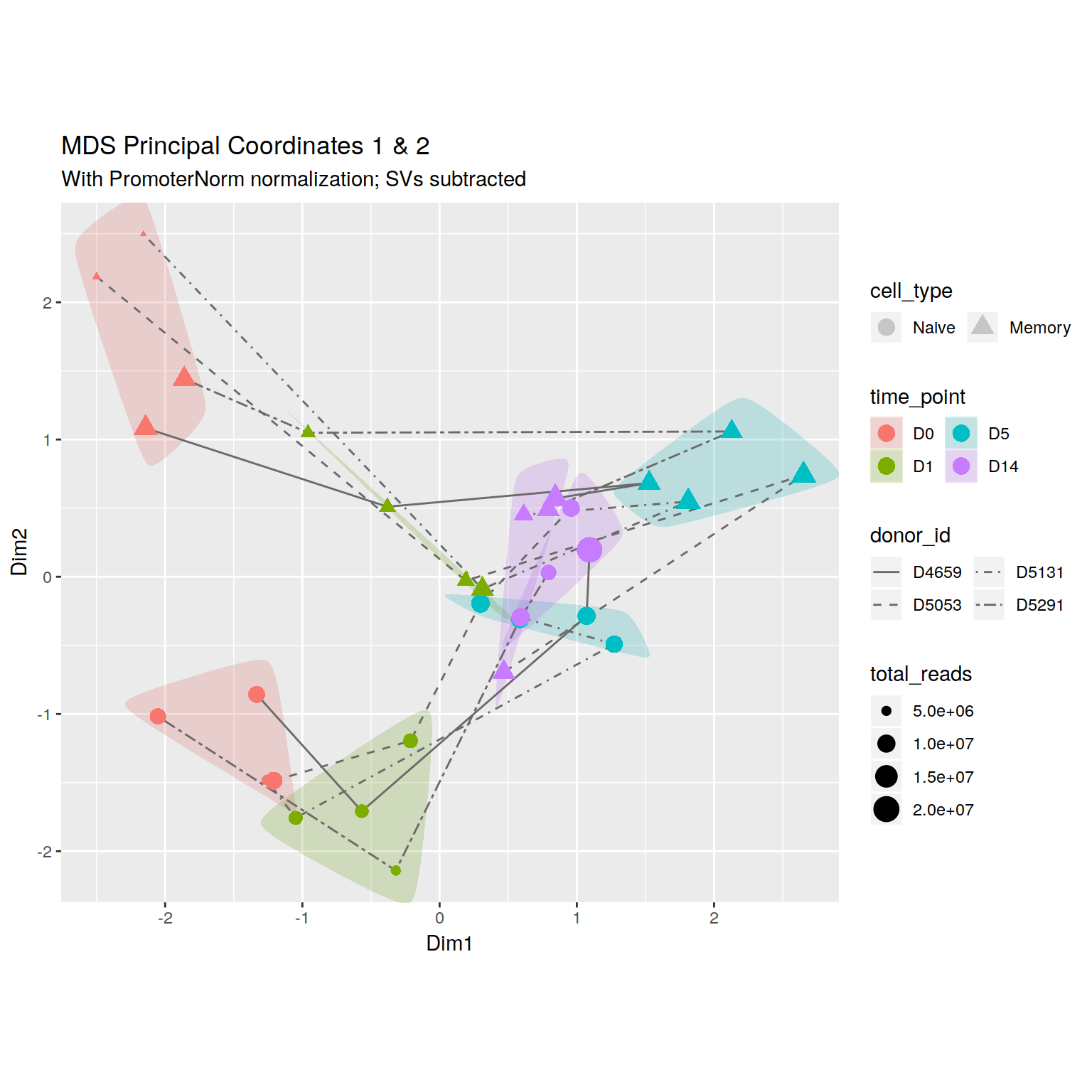
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-promoter-PCA-group-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me2-prom"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me2 promoters, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
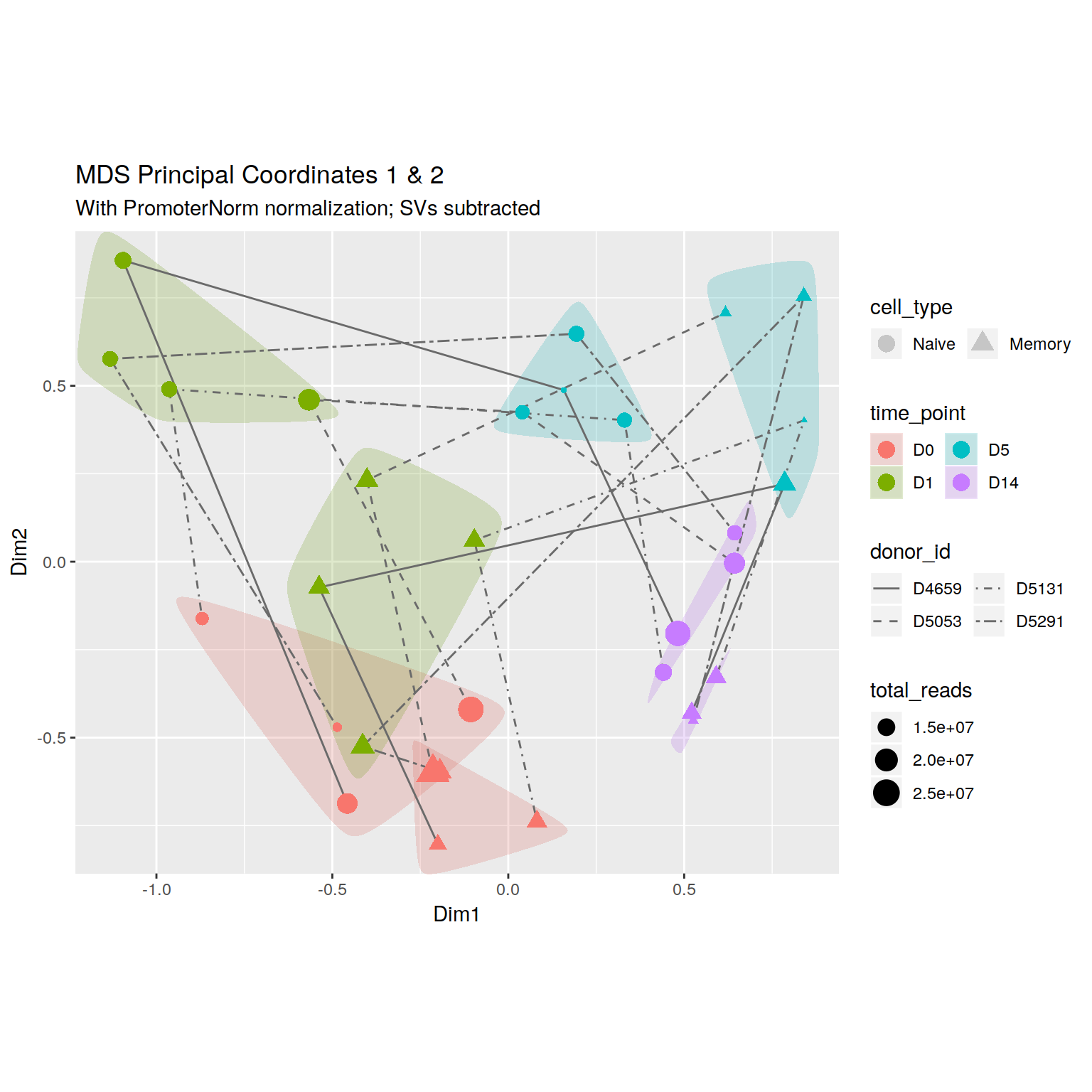
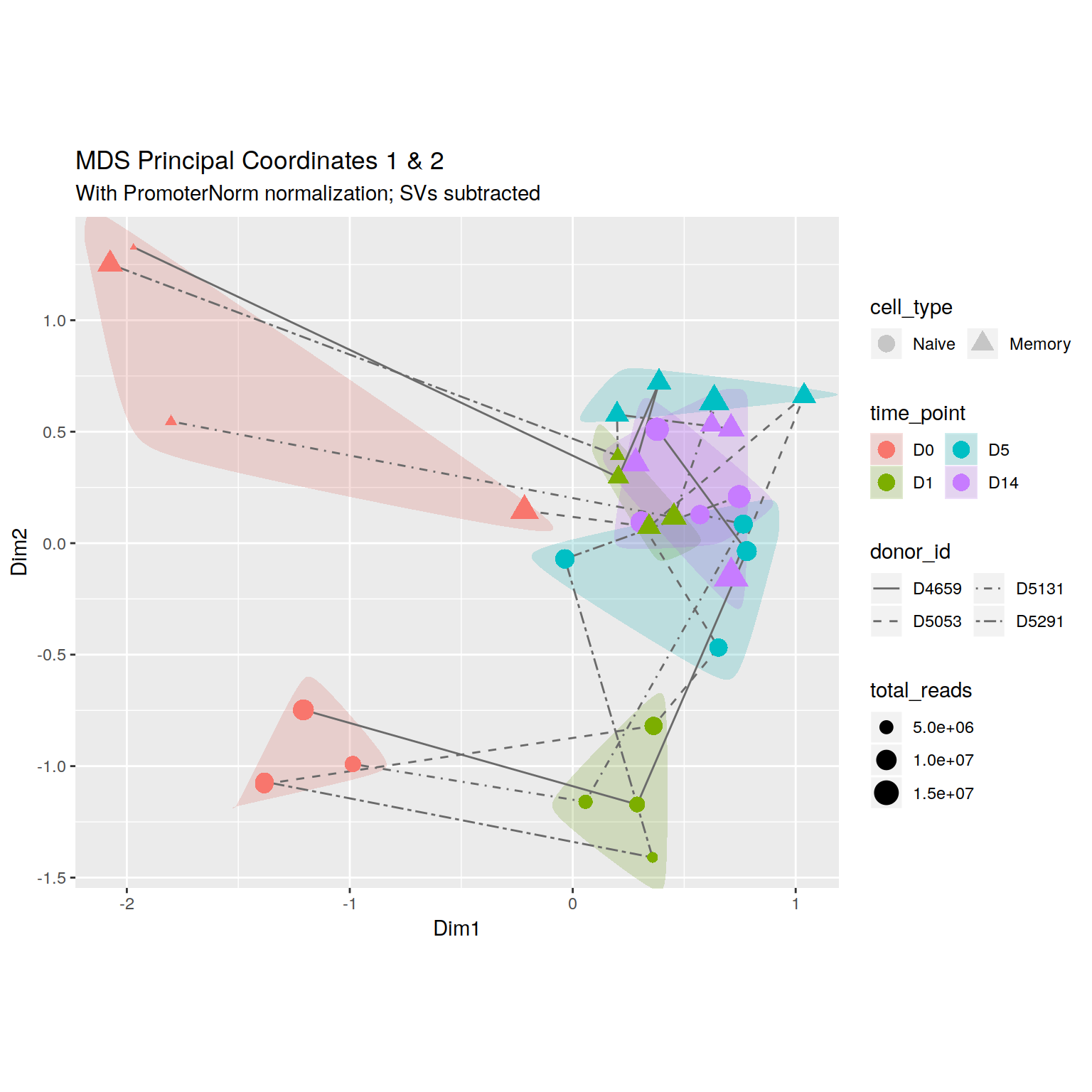
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-promoter-PCA-group-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K4me3-prom"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K4me3 promoters, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K27me3-promoter-PCA-group-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:PCoA-H3K27me3-prom"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+PCoA plot of H3K27me3 promoters, after subtracting surrogate variables
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/RNA-seq/PCA-final-23-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:RNA-PCA-group"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float table
|
|
|
+wide false
|
|
|
+sideways true
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Tabular
|
|
|
+<lyxtabular version="3" rows="6" columns="7">
|
|
|
+<features tabularvalignment="middle">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<column alignment="center" valignment="top">
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="1" alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Number of significant promoters
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="2" alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="2" alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="1" alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Est.
|
|
|
+ differentially modified promoters
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="2" alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell multicolumn="2" alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Time Point
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me2
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K27me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me2
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K4me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+H3K27me3
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Day 0
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+4553
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+927
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+6
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+9967
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+4149
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+2404
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Day 1
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+567
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+278
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+1570
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+4370
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+2145
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+6598
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Day 5
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+2313
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+139
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+490
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+9450
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+1148
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+4141
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+<row>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
|
|
|
-
|
|
|
+\begin_layout Plain Layout
|
|
|
+Day 14
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-H3K4 is correlated with higher expression, and H3K27 is correlated with
|
|
|
- lower expression genome-wide
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+0
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Flex TODO Note (inline)
|
|
|
-status open
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-Grr, gotta find these figures.
|
|
|
- Maybe in the old analysis?
|
|
|
+0
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
|
|
|
-
|
|
|
+\begin_layout Plain Layout
|
|
|
+0
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Figures showing these correlations: box/violin plots of expression distributions
|
|
|
- with every combination of peak presence/absence in promoter
|
|
|
-\end_layout
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Appropriate statistical tests showing significant differences in expected
|
|
|
- directions
|
|
|
+\begin_layout Plain Layout
|
|
|
+0
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Subsection
|
|
|
-Naive-to-memory convergence observed in H3K4 and RNA-seq data, not in H3K27me3
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+0
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Standard
|
|
|
-\begin_inset Float figure
|
|
|
-wide false
|
|
|
-sideways false
|
|
|
-status open
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+<cell alignment="center" valignment="top" topline="true" bottomline="true" leftline="true" rightline="true" usebox="none">
|
|
|
+\begin_inset Text
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
-\align center
|
|
|
-\begin_inset Graphics
|
|
|
- filename graphics/CD4-csaw/RNA-seq/PCA-final-23-CROP.png
|
|
|
- lyxscale 25
|
|
|
- width 100col%
|
|
|
- groupId colwidth-raster
|
|
|
+0
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+</cell>
|
|
|
+</row>
|
|
|
+</lyxtabular>
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
@@ -2706,11 +3650,25 @@ status open
|
|
|
\series bold
|
|
|
\begin_inset CommandInset label
|
|
|
LatexCommand label
|
|
|
-name "fig:RNA-PCA-group"
|
|
|
+name "tab:Number-signif-promoters"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+Number of differentially modified promoters between naive and memory cells
|
|
|
+ at each time point after activation.
|
|
|
+
|
|
|
+\series default
|
|
|
+This table shows both the number of differentially modified promoters detected
|
|
|
+ at a 10% FDR threshold (left half), and the total number of differentially
|
|
|
+ modified promoters as estimated using the method of
|
|
|
+\begin_inset CommandInset citation
|
|
|
+LatexCommand cite
|
|
|
+key "Phipson2013"
|
|
|
+literal "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ (right half).
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2723,12 +3681,11 @@ RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-H3K4 and RNA-seq data show clear evidence of naive convergence with memory
|
|
|
- between days 1 and 5 (Figures
|
|
|
+\begin_layout Standard
|
|
|
+Figures
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:PCoA-H3K4me2-good"
|
|
|
+reference "fig:PCoA-H3K4me2-prom"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
@@ -2738,7 +3695,7 @@ noprefix "false"
|
|
|
,
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:PCoA-H3K4me3-good"
|
|
|
+reference "fig:PCoA-H3K4me3-prom"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
@@ -2748,7 +3705,24 @@ noprefix "false"
|
|
|
, and
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:RNA-PCA-group"
|
|
|
+reference "fig:PCoA-H3K27me3-prom"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ show the patterns of variation in all 3 histone marks in the promoter regions
|
|
|
+ of the genome using principal coordinate analysis.
|
|
|
+ All 3 marks show a noticeable convergence between the naive and memory
|
|
|
+ samples at day 14, visible as an overlapping of the day 14 groups on each
|
|
|
+ plot.
|
|
|
+ This is consistent with the counts of significantly differentially modified
|
|
|
+ promoters and estimates of the total numbers of differentially modified
|
|
|
+ promoters shown in Table
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "tab:Number-signif-promoters"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
@@ -2756,37 +3730,113 @@ noprefix "false"
|
|
|
\end_inset
|
|
|
|
|
|
.
|
|
|
-\end_layout
|
|
|
+ For all histone marks, evidence of differential modification between naive
|
|
|
+ and memory samples was detected at every time point except day 14.
|
|
|
+ The day 14 convergence pattern is also present in the RNA-seq data (Figure
|
|
|
+
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:RNA-PCA-group"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Table of numbers of genes different between N & M at each time point, showing
|
|
|
- dwindling differences at later time points, consistent with convergence
|
|
|
-\end_layout
|
|
|
+\end_inset
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-Similar figure for H3K27me3 showing lack of convergence (Figure
|
|
|
+), albiet in the 2nd and 3rd principal coordinates, indicating that it is
|
|
|
+ not the most dominant pattern driving gene expression.
|
|
|
+ Taken together, the data show that promoter histone methylation for these
|
|
|
+ 3 histone marks and RNA expression for naive and memory cells are most
|
|
|
+ similar at day 14, the furthest time point after activation.
|
|
|
+ MOFA was also able to capture this day 14 convergence pattern in latent
|
|
|
+ factor 5 (Figure
|
|
|
\begin_inset CommandInset ref
|
|
|
LatexCommand ref
|
|
|
-reference "fig:PCoA-H3K27me3-good"
|
|
|
+reference "fig:mofa-lf-scatter"
|
|
|
plural "false"
|
|
|
caps "false"
|
|
|
noprefix "false"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-)
|
|
|
+), which accounts for shared variation across all 3 histone marks and the
|
|
|
+ RNA-seq data, confirming that this is a coordinated pattern across all
|
|
|
+ 4 data sets.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+This result feels shallow, somehow.
|
|
|
+ Am I oversimplifying the observation, or trivializing the amount of work
|
|
|
+ it took to get here? Shouldn't this section be more than one paragraph?
|
|
|
+ Am I just forgetting some supporting evidence that should also go here
|
|
|
+ in order to build up to the result? Or is it good that I have a simple
|
|
|
+ relatively straightforward result that doesn't take to long to explain,
|
|
|
+ and I'm just overthinking it?
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset ERT
|
|
|
+status collapsed
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+
|
|
|
+\backslash
|
|
|
+FloatBarrier
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Subsection
|
|
|
Effect of promoter coverage upstream vs downstream of TSS
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+For the figures in this section, the group labels are arbitrary, so if time
|
|
|
+ allows, it would be good to manually reorder them in a logical way, e.g.
|
|
|
+ most upstream to most downstream.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Standard
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
sideways false
|
|
|
status open
|
|
|
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+These really need to be sub-figures A, B, and C.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
@@ -2812,7 +3862,46 @@ name "fig:H3K4me2-neighborhood-clusters"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+K-means clustering of promoter H3K4me2 relative coverage depth in naive
|
|
|
+ day 0 samples.
|
|
|
+
|
|
|
+\series default
|
|
|
+H3K4me2 ChIP-seq reads were binned into 500-bp windows tiled across each
|
|
|
+ promoter from 5
|
|
|
+\begin_inset space ~
|
|
|
+\end_inset
|
|
|
+
|
|
|
+kbp upstream to 5
|
|
|
+\begin_inset space ~
|
|
|
+\end_inset
|
|
|
+
|
|
|
+kbp downstream, and the logCPM values were normalized within each promoter
|
|
|
+ to an average of 0, yielding relative coverage depths.
|
|
|
+ These were then grouped using K-means clustering with
|
|
|
+\begin_inset Formula $K=6$
|
|
|
+\end_inset
|
|
|
+
|
|
|
+, and the average bin values were plotted for each cluster.
|
|
|
+ The
|
|
|
+\begin_inset Formula $x$
|
|
|
+\end_inset
|
|
|
+
|
|
|
+-axis is the genomic coordinate of each bin relative to the the transcription
|
|
|
+ start site, and the
|
|
|
+\begin_inset Formula $y$
|
|
|
+\end_inset
|
|
|
+
|
|
|
+-axis is the mean relative coverage depth of that bin across all promoters
|
|
|
+ in the cluster.
|
|
|
+ Each line represents the average
|
|
|
+\begin_inset Quotes eld
|
|
|
+\end_inset
|
|
|
+
|
|
|
+shape
|
|
|
+\begin_inset Quotes erd
|
|
|
+\end_inset
|
|
|
+
|
|
|
+ of the promoter coverage for promoters in that cluster.
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2856,7 +3945,23 @@ name "fig:H3K4me2-neighborhood-pca"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+PCA of promoter H3K4me2 relative coverage depth in naive day 0 samples,
|
|
|
+ colored by K-means cluster membership.
|
|
|
+
|
|
|
+\series default
|
|
|
+PCA was performed on the same relative promoter coverage values used for
|
|
|
+ K-means clustering in Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:H3K4me2-neighborhood-clusters"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+, and the first two principal components were plotted, coloring each point
|
|
|
+ by its K-means cluster identity.
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -2900,7 +4005,21 @@ name "fig:H3K4me2-neighborhood-expression"
|
|
|
|
|
|
\end_inset
|
|
|
|
|
|
-RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+Gene expression distributions grouped by H3K4me2 promoter coverage K-means
|
|
|
+ clustering.
|
|
|
+
|
|
|
+\series default
|
|
|
+For each of the clusters in Figure
|
|
|
+\begin_inset CommandInset ref
|
|
|
+LatexCommand ref
|
|
|
+reference "fig:H3K4me2-neighborhood-clusters"
|
|
|
+plural "false"
|
|
|
+caps "false"
|
|
|
+noprefix "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -3099,6 +4218,16 @@ Try to boil it down to 3 main messages to get across
|
|
|
\end_layout
|
|
|
|
|
|
\begin_deeper
|
|
|
+\begin_layout Itemize
|
|
|
+Naive-to-memory convergence in certain data sets but not others, implies
|
|
|
+ which marks are involved in memory differentiation
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_deeper
|
|
|
+\begin_layout Subsection
|
|
|
+Effective promoter radius
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
"Promoter radius" is not constant and must be defined empirically for a
|
|
|
given data set.
|
|
|
@@ -3106,15 +4235,15 @@ Try to boil it down to 3 main messages to get across
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Itemize
|
|
|
-Naive-to-memory convergence in certain data sets but not others, implies
|
|
|
- which marks are involved in memory differentiation
|
|
|
+Further study required to demonstarte functional consequences of effective
|
|
|
+ promoter radius (e.g.
|
|
|
+ show diminished association with gene expression outside radius)
|
|
|
\end_layout
|
|
|
|
|
|
-\begin_layout Itemize
|
|
|
-TSS positional coverage, hints of something interesting but no clear conclusions
|
|
|
+\begin_layout Subsection
|
|
|
+Convergence
|
|
|
\end_layout
|
|
|
|
|
|
-\end_deeper
|
|
|
\begin_layout Standard
|
|
|
\begin_inset Float figure
|
|
|
wide false
|
|
|
@@ -3178,8 +4307,16 @@ H3K27me3, canonically regarded as a deactivating mark, seems to have a more
|
|
|
\end_layout
|
|
|
|
|
|
\end_deeper
|
|
|
+\begin_layout Subsection
|
|
|
+Positional
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
-TSS positional coverage
|
|
|
+TSS positional coverage, hints of something interesting but no clear conclusions
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Subsection
|
|
|
+Workflow
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|
|
|
@@ -3236,6 +4373,10 @@ Decision-making based on trying every option and running the workflow downstream
|
|
|
\end_layout
|
|
|
|
|
|
\end_deeper
|
|
|
+\begin_layout Subsection
|
|
|
+Data quality issues limit conclusions
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Chapter
|
|
|
Improving array-based analyses of transplant rejection by optimizing data
|
|
|
preprocessing
|
|
|
@@ -6577,7 +7718,7 @@ noprefix "false"
|
|
|
\begin_inset Float table
|
|
|
wide false
|
|
|
sideways false
|
|
|
-status collapsed
|
|
|
+status open
|
|
|
|
|
|
\begin_layout Plain Layout
|
|
|
\align center
|
|
|
@@ -6811,7 +7952,15 @@ noprefix "false"
|
|
|
\end_inset
|
|
|
|
|
|
, the table shows the number of probes estimated to be differentially methylated
|
|
|
- between TX and the other 3 transplant statuses.
|
|
|
+ between TX and the other 3 transplant statuses using the method of
|
|
|
+\begin_inset CommandInset citation
|
|
|
+LatexCommand cite
|
|
|
+key "Phipson2013"
|
|
|
+literal "false"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+.
|
|
|
\end_layout
|
|
|
|
|
|
\end_inset
|
|
|
@@ -9715,6 +10864,20 @@ Check bib entry formatting & sort order
|
|
|
\end_inset
|
|
|
|
|
|
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Flex TODO Note (inline)
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+Check in-text citation format.
|
|
|
+ Probably don't just want [1], [2], etc.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
\end_layout
|
|
|
|
|
|
\begin_layout Standard
|