|
|
@@ -558,6 +558,7 @@ status open
|
|
|
\align center
|
|
|
\begin_inset Graphics
|
|
|
filename graphics/CD4-csaw/rulegraphs/rulegraph-all.pdf
|
|
|
+ lyxscale 50
|
|
|
width 100theight%
|
|
|
|
|
|
\end_inset
|
|
|
@@ -1512,6 +1513,7 @@ Comparison of quantification between Salmon with and without Shoal for identical
|
|
|
Ultimately selected shoal as quantification, Ensembl as annotation.
|
|
|
Why? Running downstream analyses with all quant methods and both annotations
|
|
|
showed very little practical difference, so choice was not terribly important.
|
|
|
+ Prefer shoal due to theoretical advantages.
|
|
|
To note in discussion: reproducible workflow made it easy to do this, enabling
|
|
|
an informed decision.
|
|
|
\end_layout
|
|
|
@@ -2648,6 +2650,270 @@ noprefix "false"
|
|
|
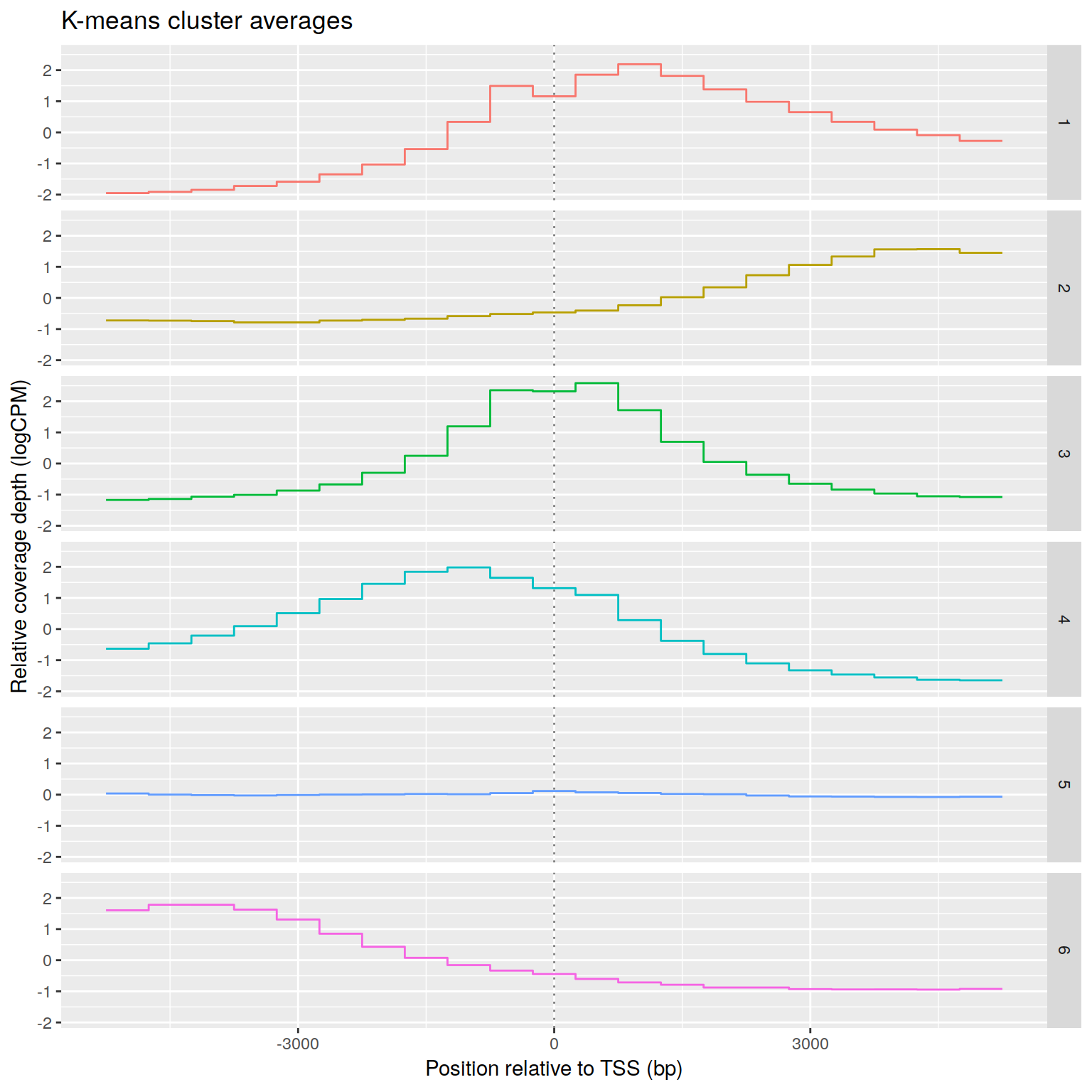
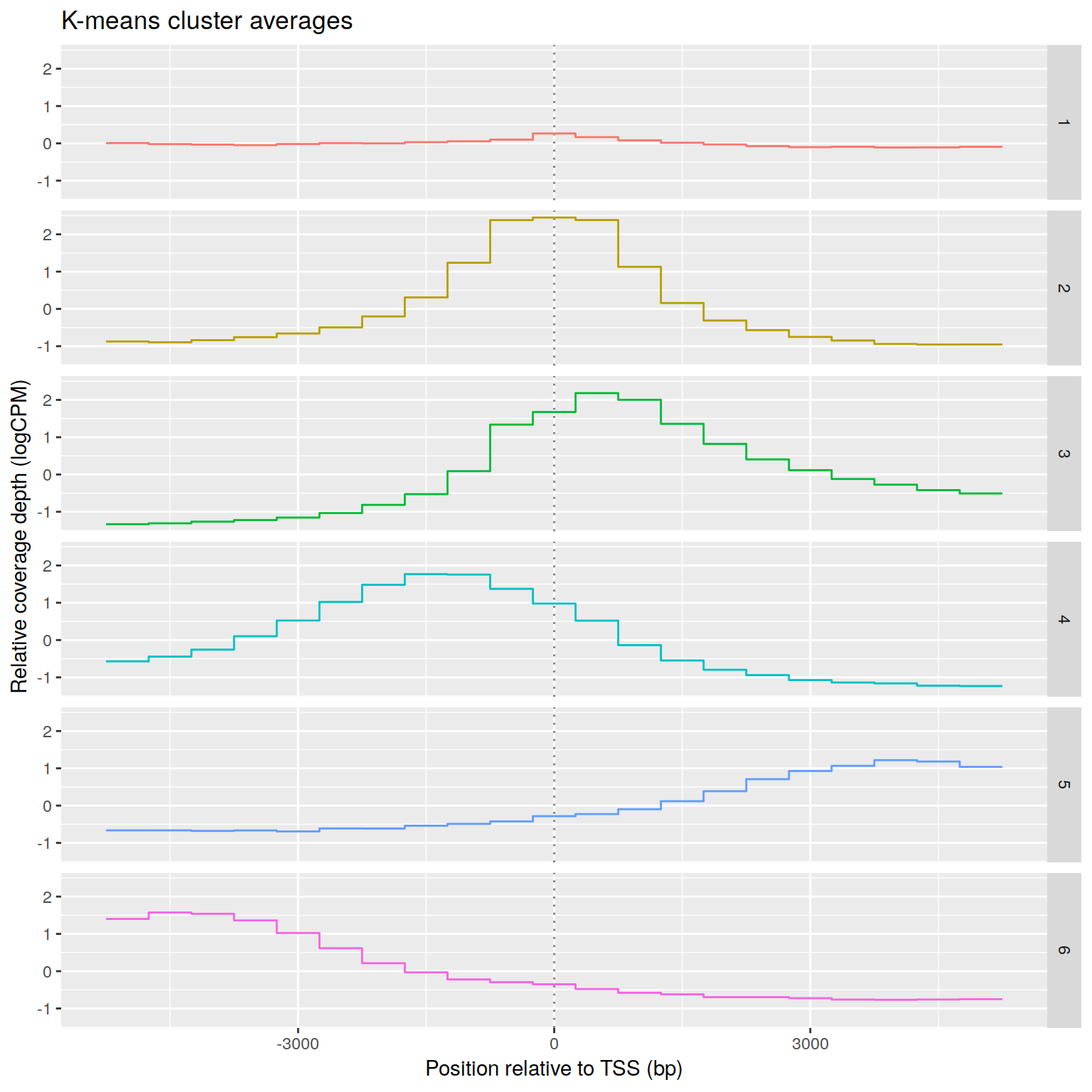
Effect of promoter coverage upstream vs downstream of TSS
|
|
|
\end_layout
|
|
|
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-neighborhood-clusters-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me2-neighborhood-clusters"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
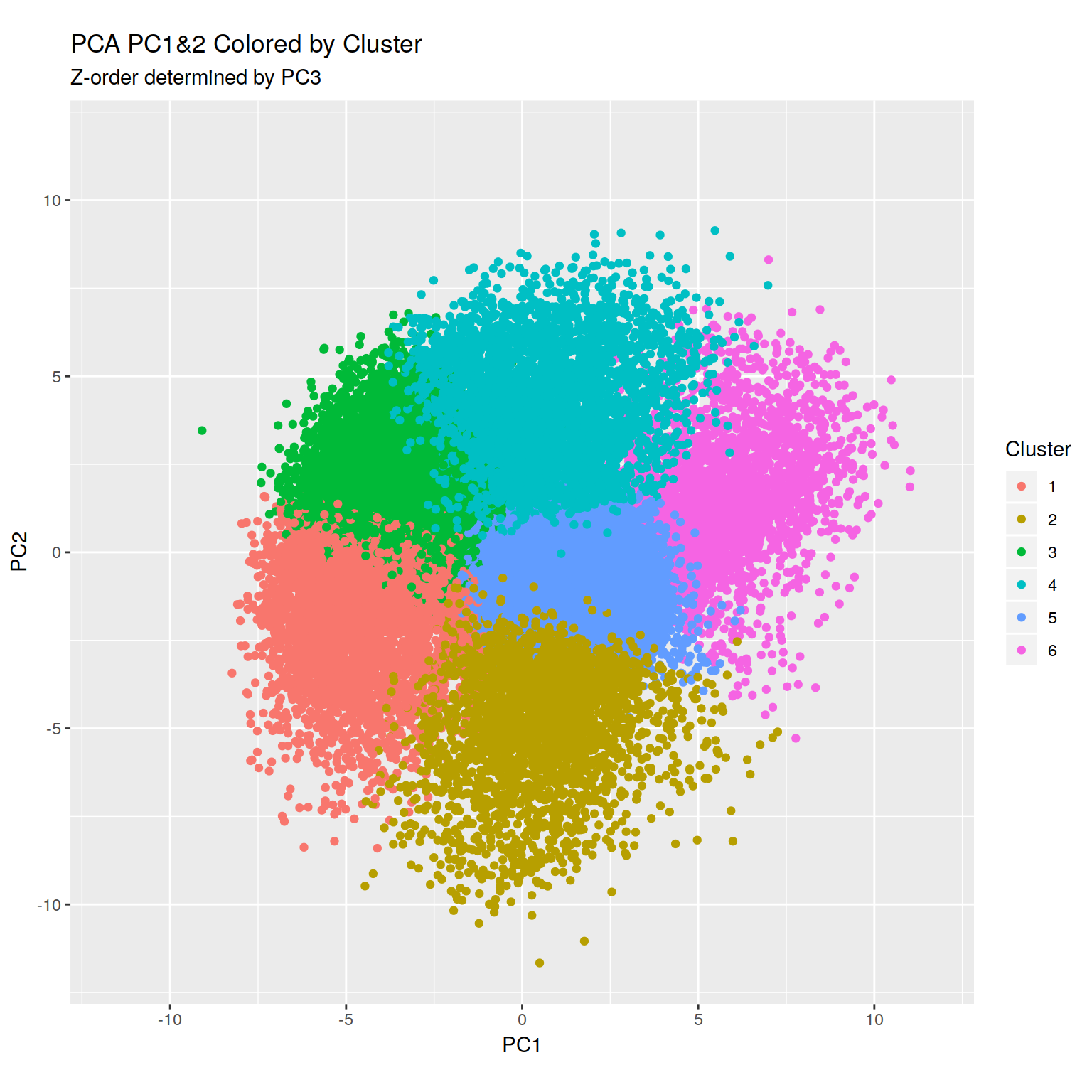
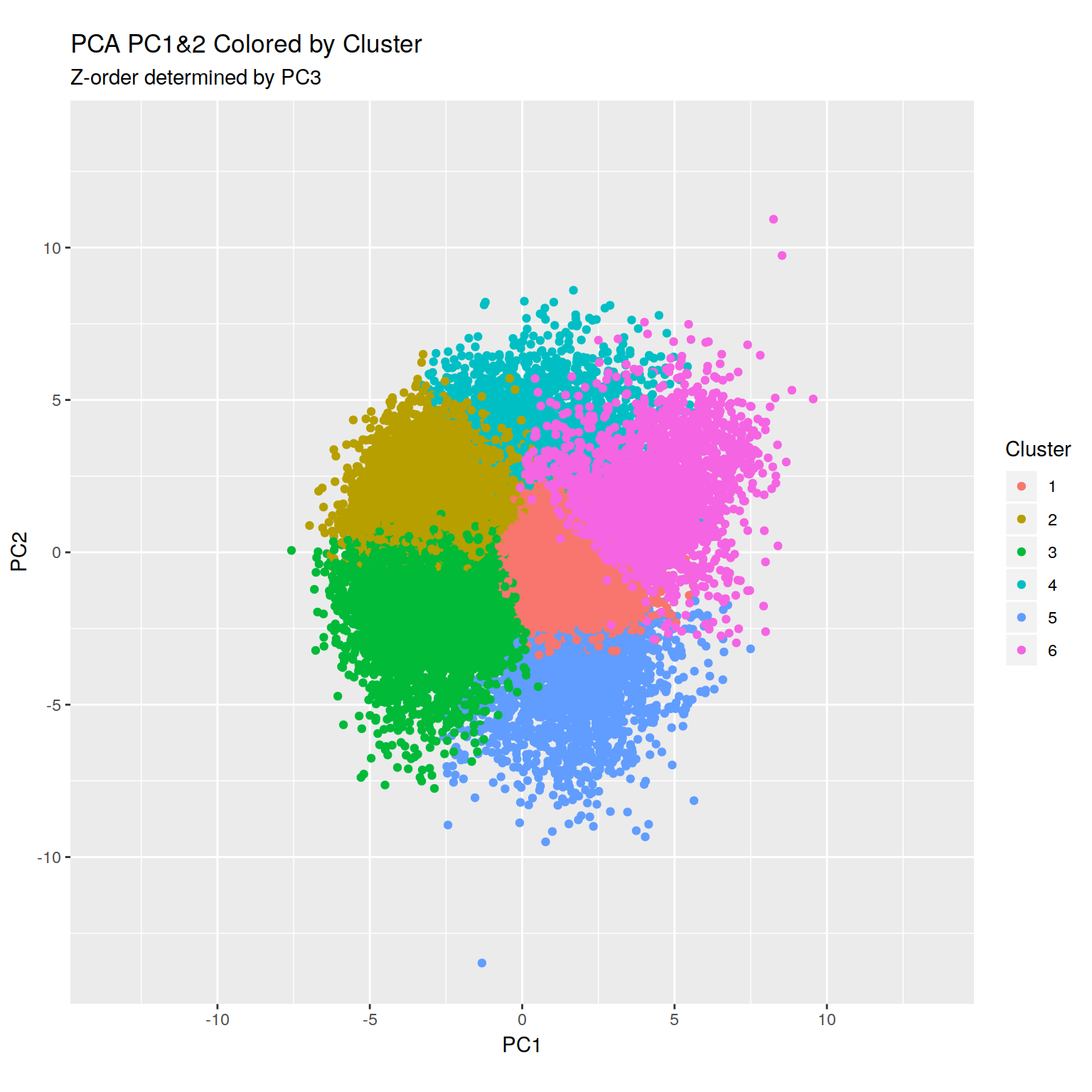
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-neighborhood-PCA-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me2-neighborhood-pca"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
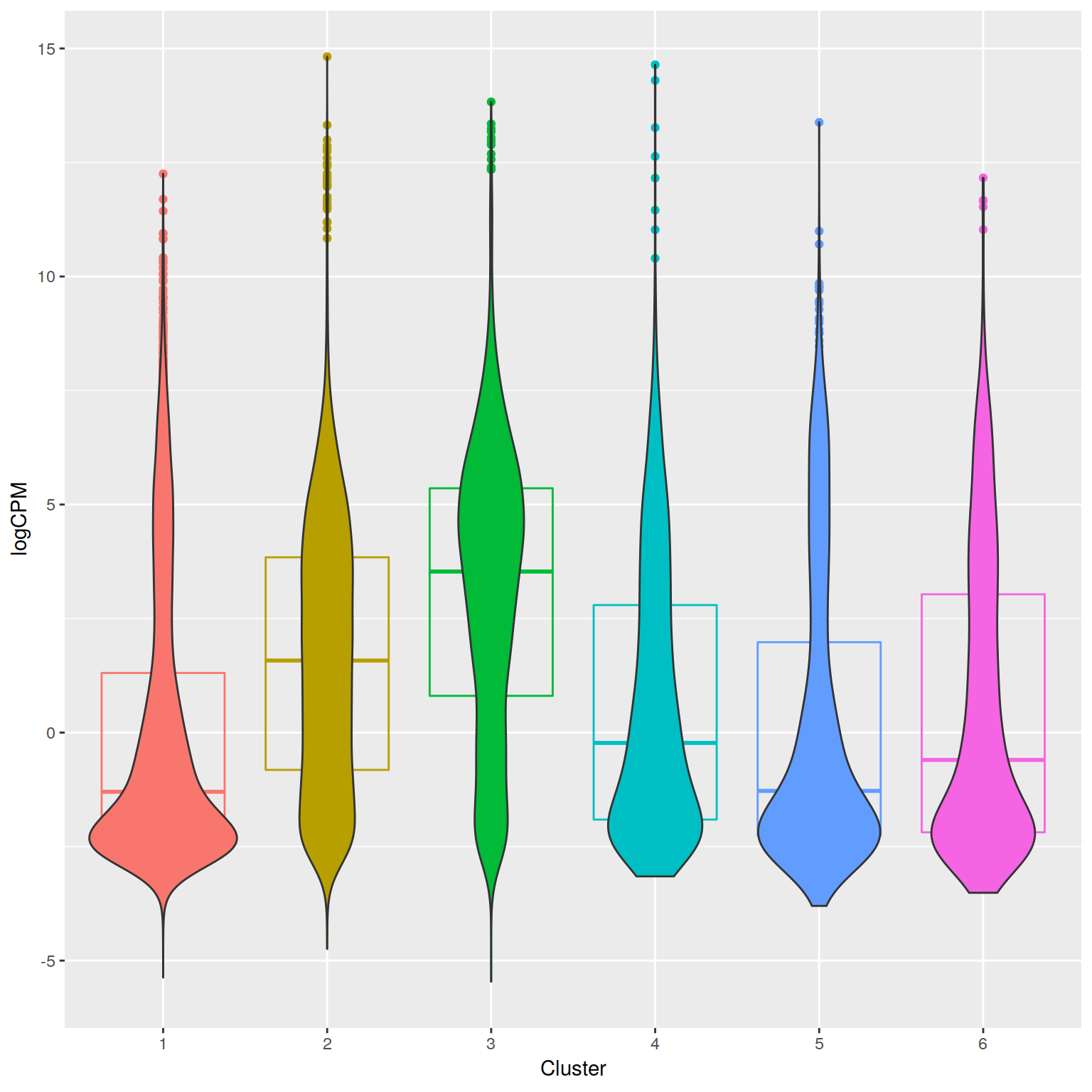
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me2-neighborhood-expression-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me2-neighborhood-expression"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
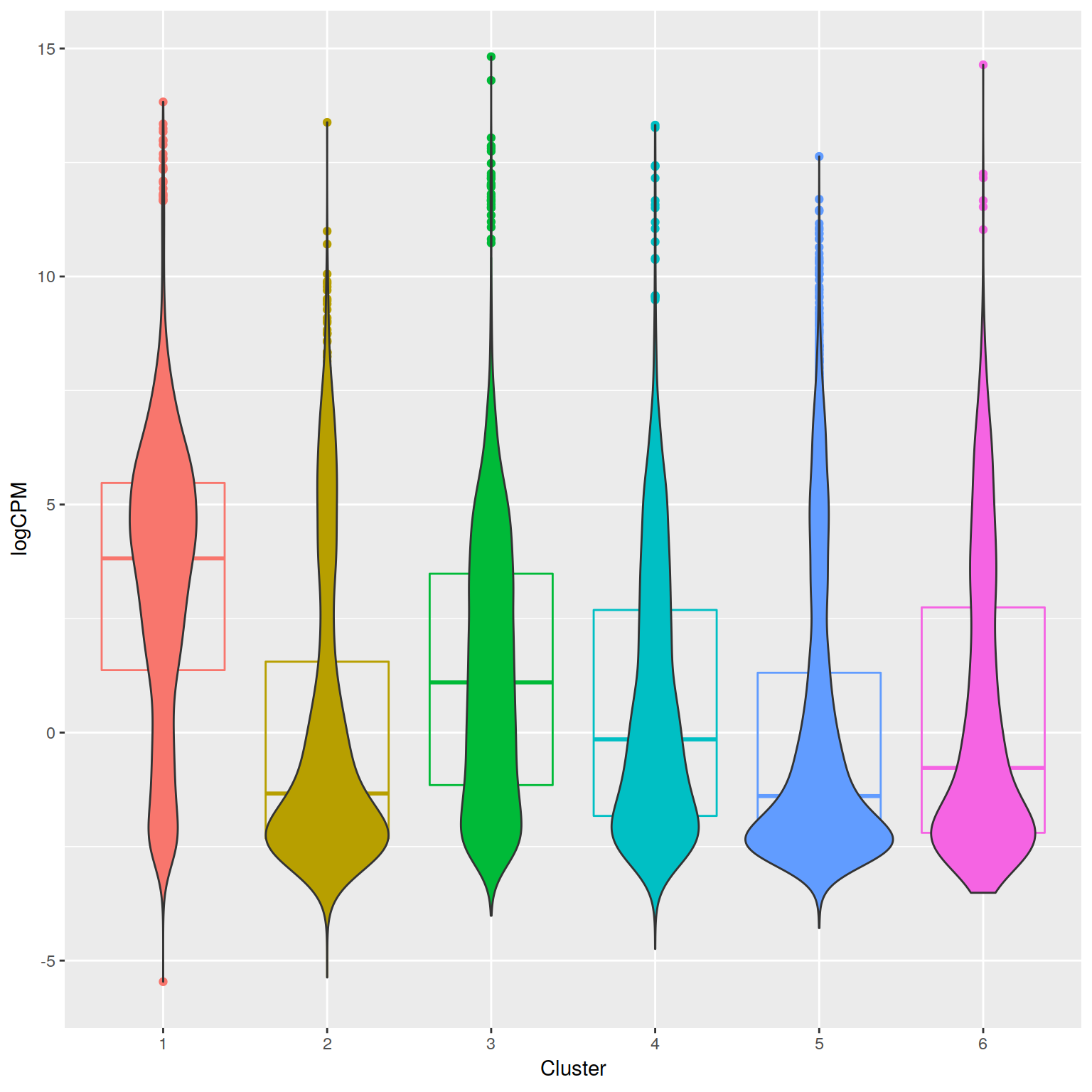
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-neighborhood-clusters-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me3-neighborhood-clusters"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-neighborhood-PCA-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me3-neighborhood-pca-1"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Standard
|
|
|
+\begin_inset Float figure
|
|
|
+wide false
|
|
|
+sideways false
|
|
|
+status open
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\align center
|
|
|
+\begin_inset Graphics
|
|
|
+ filename graphics/CD4-csaw/ChIP-seq/H3K4me3-neighborhood-expression-CROP.png
|
|
|
+ lyxscale 25
|
|
|
+ width 100col%
|
|
|
+ groupId colwidth-raster
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+\begin_inset Caption Standard
|
|
|
+
|
|
|
+\begin_layout Plain Layout
|
|
|
+
|
|
|
+\series bold
|
|
|
+\begin_inset CommandInset label
|
|
|
+LatexCommand label
|
|
|
+name "fig:H3K4me3-neighborhood-expression-1"
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+RNA-seq PCoA showing principal coordiantes 2 and 3.
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
+\end_inset
|
|
|
+
|
|
|
+
|
|
|
+\end_layout
|
|
|
+
|
|
|
\begin_layout Itemize
|
|
|
H3K4me peaks seem to correlate with increased expression as long as they
|
|
|
are anywhere near the TSS
|