|
|
@@ -160,12 +160,6 @@ Compared to naïve cells, memory cells:
|
|
|
naïve cells
|
|
|
* evolve over time to respond even more strongly to their antigen
|
|
|
|
|
|
-:::
|
|
|
-
|
|
|
-## Memory cells are a problem for immune suppression
|
|
|
-
|
|
|
-\large
|
|
|
-
|
|
|
Result:
|
|
|
|
|
|
\normalsize
|
|
|
@@ -175,22 +169,23 @@ Result:
|
|
|
* Dosage cannot be increased indefinitely without compromising the
|
|
|
immune system's ability to fight infection
|
|
|
|
|
|
+:::
|
|
|
+
|
|
|
## We need a better understanding of immune memory
|
|
|
|
|
|
-* Cell surface markers of naïve and memory $\mathsf{CD4}^{+}$ T-cells
|
|
|
- are fairly well-characterized
|
|
|
-* But internal mechanisms that allow memory cells to respond
|
|
|
- differently to the same stimulus (antigen presentation) are not
|
|
|
- well-understood
|
|
|
+* Cell surface markers fairly well-characterized
|
|
|
+
|
|
|
+* But internal mechanisms poorly understood
|
|
|
|
|
|
. . .
|
|
|
|
|
|
-* A reasonable hypothesis is that some of these mechanisms are
|
|
|
- epigenetic: using histone marks or DNA methylation to regulate the
|
|
|
- expression of certain genes
|
|
|
-* We can test this hypothesis by measuring gene expression (using
|
|
|
- RNA-seq) and histone methylation (using ChIP-seq) in naïve and
|
|
|
- memory T-cells before and after activation
|
|
|
+\vfill
|
|
|
+
|
|
|
+\large
|
|
|
+
|
|
|
+**Hypothesis:** Epigenetic regulation of gene expression through
|
|
|
+histone modification is involved in $\mathsf{CD4}^{+}$ T-cell
|
|
|
+activation and memory.
|
|
|
|
|
|
## Experimental design
|
|
|
|
|
|
@@ -205,6 +200,27 @@ Result:
|
|
|
Data generated by Sarah Lamere, published in GEO as
|
|
|
[GSE73214](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73214)
|
|
|
|
|
|
+## Time points capture phases of immune response
|
|
|
+
|
|
|
+\centering
|
|
|
+
|
|
|
+<!-- { height=75% } -->
|
|
|
+
|
|
|
+## Why study these histone marks?
|
|
|
+
|
|
|
+::: incremental
|
|
|
+
|
|
|
+* **H3K4me3:** "activating" mark associated with active transcription
|
|
|
+
|
|
|
+* **H3K4me2:** Correlated with H3K4me3, hypothesized as a "poised" state
|
|
|
+
|
|
|
+* **H3K27me3:** "repressive" mark associated with inactive
|
|
|
+
|
|
|
+* All 3 involved in T-cell differentiation, but activation dynamics
|
|
|
+ unexplored
|
|
|
+
|
|
|
+:::
|
|
|
+
|
|
|
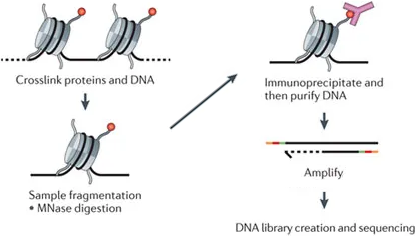
## ChIP-seq sequences DNA bound to marked histones[^chipseq]
|
|
|
|
|
|
\centering
|
|
|
@@ -213,10 +229,6 @@ Data generated by Sarah Lamere, published in GEO as
|
|
|
|
|
|
[^chipseq]: [Furey. "ChIP-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions". In: Nature Reviews Genetics (2012)](http://www.nature.com/articles/nrg3306)
|
|
|
|
|
|
-## H3K4me2, H3K4me3, H3K27me3
|
|
|
-
|
|
|
-Why? <!-- TODO -->
|
|
|
-
|
|
|
## A few intermediate analysis steps are required
|
|
|
|
|
|

|