|
|
@@ -1,7 +1,7 @@
|
|
|
<ul>
|
|
|
<li><a href="delox.R.html"><code>delox.R.html</code></a>: The R code for <a href="https://academic.oup.com/nar/article/40/3/e24/1128321/Illumina-mate-paired-DNA-sequencing-library">Deloxer</a></li>
|
|
|
<li><a href="depth-consistency.pdf"><code>depth-consistency.pdf</code></a>: Visualization of consistency of coverage depth between 2 Exome sequencing replicates</li>
|
|
|
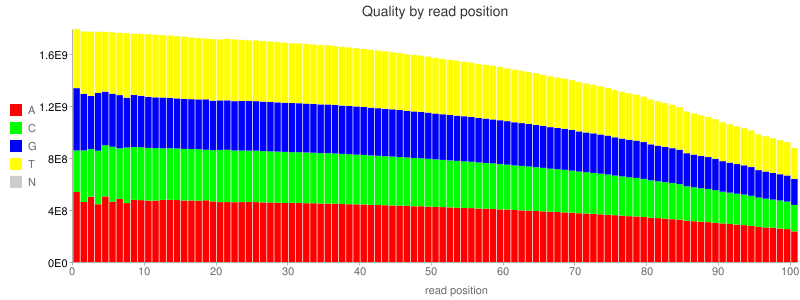
-<li><a href="illumina-qc.html"><code>illumina-qc.html</code></a>: QC chart indicating quality score dropoff along read length for exome sequencing data set.</li>
|
|
|
+<li><a href="illumina-qc.png"><code>illumina-qc.html</code></a>: QC chart indicating quality score dropoff along read length for exome sequencing data set.</li>
|
|
|
<li><a href="ipi-results-small.txt"><code>ipi-results-small.txt</code></a>: A sample of output showing alignment of <em>de novo</em> assembled black rhino and white rhino transcript sequences to human proteins. This was used to find differences between the two rhino species in otherwise conserved regions.</li>
|
|
|
<li><a href="mirna-results.html"><code>mirna-results.html</code></a>: Example output from miRNA binding site predictions</li>
|
|
|
<li><a href="neartarget.pdf"><code>neartarget.pdf</code></a>: Visualization of mean coverage depth in exome sequencing data in the regions flanking the probe targets.</li>
|